Macromolecular Modeling Platform
Staff

Laurent LE CORRE
Operational Engineer, IE (CNRS)
Email: laurent.le-corre@parisdescartes.fr
Location
UMR 8601
Room R150
Equipments
- 1 Precision 7920 Desktop workstation, 2x Intel ® Xeon ® Gold 6248R CPU@ 3.00GHz, 48 cores, 96 threads (2022)
- 1 Precision 7820 Desktop workstation, 2x Intel ® Xeon ® Gold 5118 CPU@2.30 GHz-3.2 GHz Turbo, 24 cores, 48 threads (2019)
- 1 Precision 7810 Desktop workstation, 2x Intel ® Xeon ® CPU E5 2640 v3 @2.60 GHz-3.40 GHz, 16 cores, 32 threads (2016)
- Commercial software : Biovia Discovery Studio 2021
The Interdisciplinary Macromolecular Modeling Platform located into the LCBPT (Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques) offers its expertise to answer structural and functional questions on molecules or complexes of biological interest.
Activities range from visualization of experimental three-dimensional structures and their mutants, to more complex questions such as protein 3D model generation, ligand/receptor interaction (docking, molecular dynamics, simulations), structure / activity studies, virtual screening. The board is open to a collaborative type search.
Recent in silico screening applications:
Over the last 5 years, at least 15 in silico screening (mainly from our in-house library) has been performed mainly to identify new small molecules able to modulate original biological target activity implicated in several therapeutic areas including infections, cancers, mood diseases. Among the most interesting hits, we have notably identified 2 original hits targeting the RED-SMU1 complex with antiviral activity and led to medicinal chemistry “hit to lead” program (collaboration Institut Pasteur, the Institut de Biologie Structurale at Grenoble, the Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques at Univ Paris-Descartes, and the PRABI at Univ Claude-Bernard).
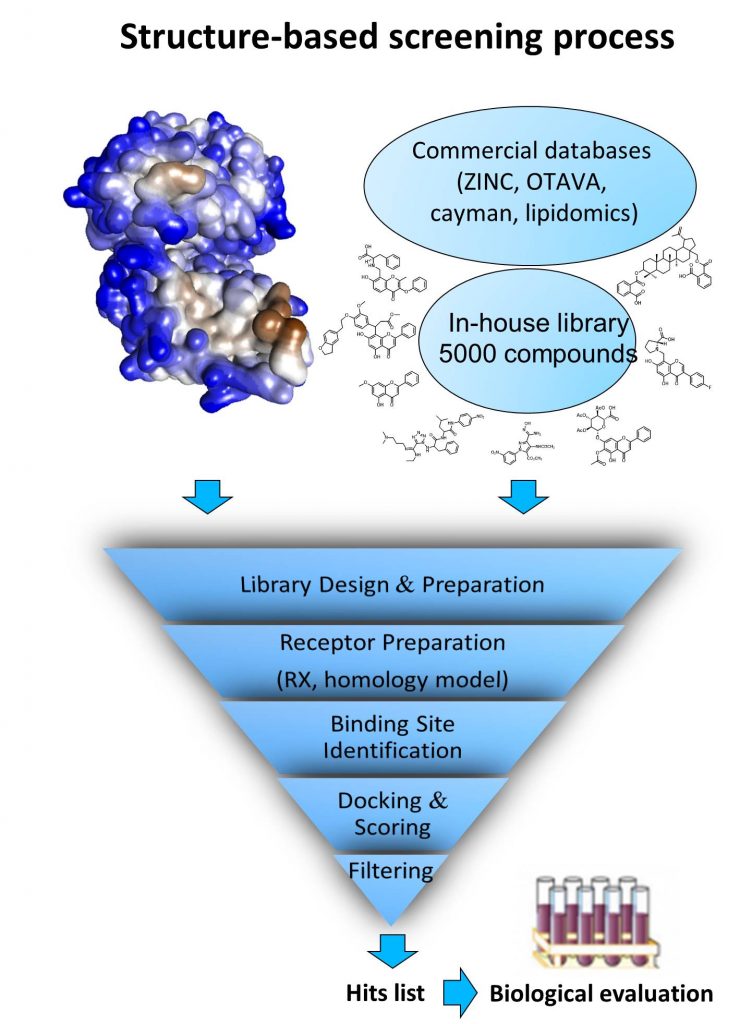
Structure/activity studies:
The last aspect of our work remains the optimization of chemical hit. By working in closed collaboration with chemists and biologists, we performed docking experiments and proposed various modifications to increase compounds potency and selectivity.
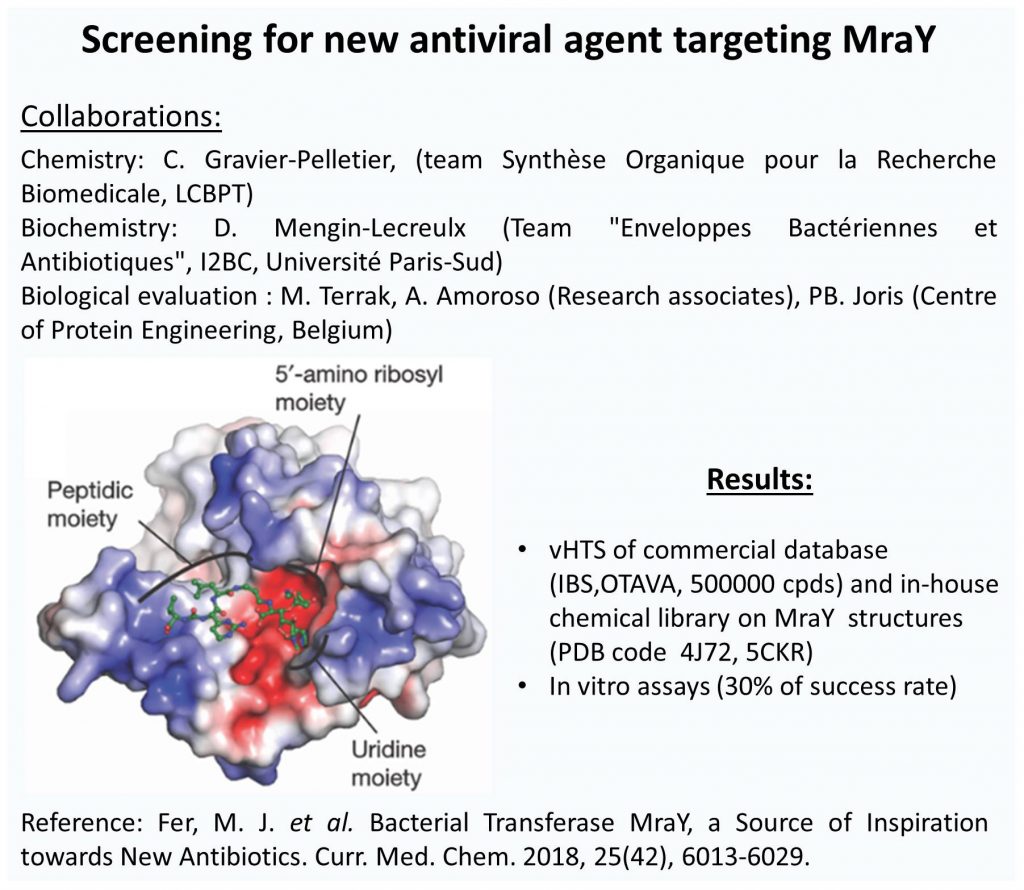
Structural aspects:
Using molecular modeling approaches, we have deciphered the binding mode of substrates such as lipid IIby the peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae in closedcollaboration with structural expert (Institut de Biologie Structurale at Grenoble).
Another part of our program consist in the identification of putative substrate of characterized protein such as CYP450 and the impact of various mutation. To reach this goal we used both virtual mutagenesis and natural databank compound screening.
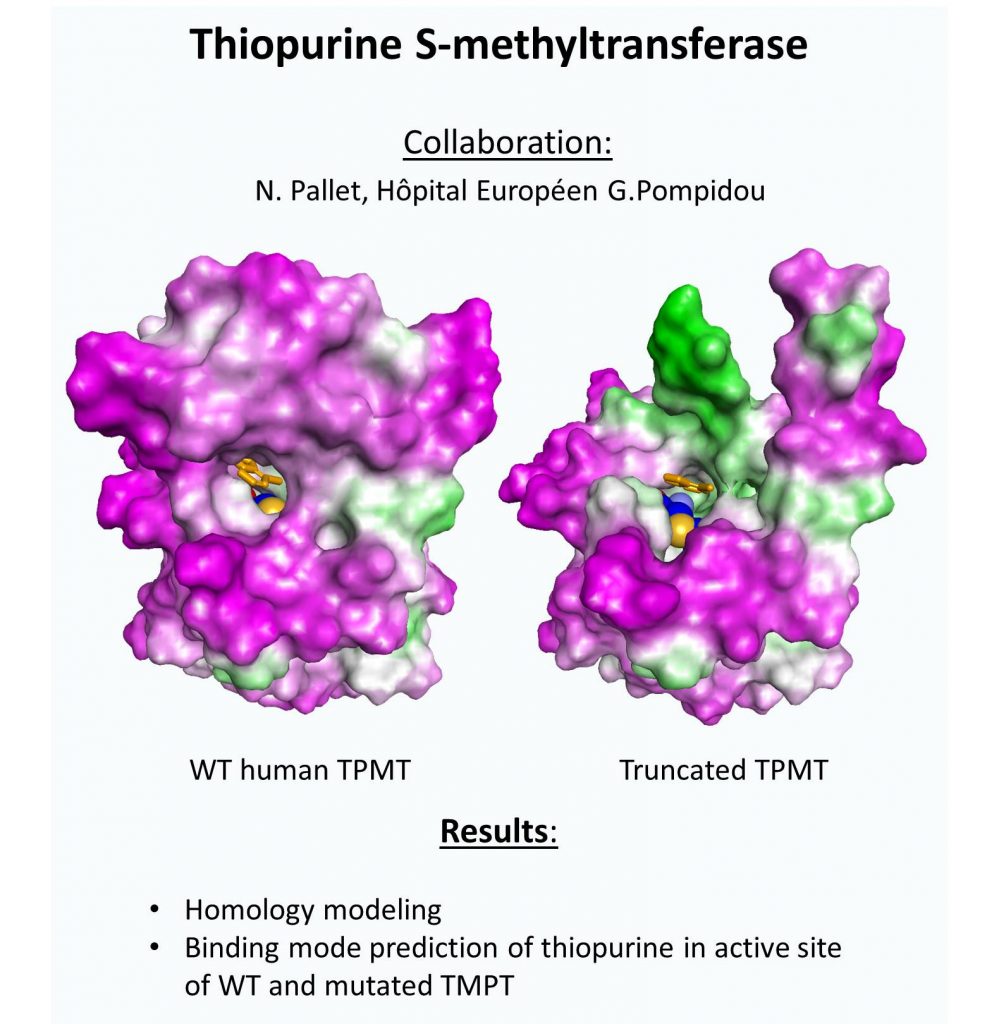
Macromolecular Modeling Platform
Staff

Laboratoire de Chimie et Biochimie Pharmacologique et Toxicologique (LCBPT),
UMR8601,
Room R150
Equipments
- 1 Precision 7920 Desktop workstation, 2x Intel® Xeon® Gold 6248R CPU @ 3.00GHz 2.99 GHz, 48 cores, 96 threads (2022)
- 1 Precision 7820 Desktop workstation, 2x Intel ® Xeon ® Gold 5118 CPU@2.30 GHz-3.2 GHz Turbo, 24 cores, 48 threads (2019)
- 1 Precision 7810 Desktop workstation, 2x Intel ® Xeon ® CPU E5 2640 v3 @2.60 GHz-3.40 GHz, 16 cores, 32 threads (2016)
- Commercial software : Biovia Discovery Studio 2022
Description
The Interdisciplinary Macromolecular Modeling Platform located into the LCBPT (Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques) offers its expertise to answer structural and functional questions on molecules or complexes of biological interest.
Activities range from visualization of experimental three-dimensional structures and their mutants, to more complex questions such as protein 3D model generation, ligand/receptor interaction (docking, molecular dynamics, simulations), structure / activity studies, virtual screening. The board is open to a collaborative type search.
Recent in silico screening applications:
Over the last 5 years, at least 15 in silico screening (mainly from our in-house library) has been performed mainly to identify new small molecules able to modulate original biological target activity implicated in several therapeutic areas including infections, cancers, mood diseases. Among the most interesting hits, we have notably identified 2 original hits targeting the RED-SMU1 complex with antiviral activity and led to medicinal chemistry “hit to lead” program (collaboration Institut Pasteur, the Institut de Biologie Structurale at Grenoble, the Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques at Univ Paris-Descartes, and the PRABI at Univ Claude-Bernard).

Structure/activity studies:
The last aspect of our work remains the optimization of chemical hit. By working in closed collaboration with chemists and biologists, we performed docking experiments and proposed various modifications to increase compounds potency and selectivity.

Structural aspects:
Using molecular modeling approaches, we have deciphered the binding mode of substrates such as lipid IIby the peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae in closedcollaboration with structural expert (Institut de Biologie Structurale at Grenoble).
Another part of our program consist in the identification of putative substrate of characterized protein such as CYP450 and the impact of various mutation. To reach this goal we used both virtual mutagenesis and natural databank compound screening.

Main publications
- Coelho, D.; Le Corre, L.; Bartosik, K.; Iannazzo, L.; Braud, E.; Etheve-Quelquejeu, M. “Synthesis of Bisubstrate Analogues for RNA Methylation Studies using two Transition Metal-Catalyzed Reactions” Chem. Eur. J. 2023, e202301134.
- Le Corre, L.; Padovani, D. “Mechanism‑based and computational modeling of hydrogen sulfide biogenesis inhibition: interfacial inhibition.” Scientific Reports 2023, 13:7287. https://doi.org/10.1038/s41598-023-34405-3
- Galardon, E.; Mathas, N.; Padovani, D.; Le Corre, L.; Poncet, G.; Dairou, J. “Persulfidation of DJ-1: Mechanism and Consequences.” Biomolecules 2023, 13, 27. https://doi.org/10.3390/biom13010027
- Nouri, K.; Pietrancosta, N.; Le Corre, L.; Dansette, P.M.; Mansuy,D.; Boucher, J.-L. “Human Orphan Cytochrome P450 2U1 Catalyzes the ω-Hydroxylation of Leukotriene B4.” Int. J. Mol. Sci. 2022, 23, 14615. https://doi.org/10.3390/ijms232314615
- M. Oliver, L, Le Corre, M. Poinsot, M, Bosco, H. Wan, A. Amoroso, B. Joris, A. Bouhss, S. Calvet-Vitale, C. Gravier-Pelletier “A Sub-Micromolar MraYAA Inhibitor with an Aminoribosyl Uridine Structure and a (S,S)-Tartaric Diamide: Synthesis, Biological Evaluation and Molecular Modeling.” Molecules 2022, 27(6), 1769. https://doi.org/10.3390/molecules27061769
- Pujol C, Legrand A, Parodi L, Boucher, J.-L.; Le Corre, L. et al. “Implication of folate deficiency in CYP2U1 loss of function.” J Exp Med. 2021; 218(11):e20210846. https://doi.org/10.1084/jem.20210846
- Oliver, M.; Le Corre, L.; Poinsot, M.; Bosco, M.; Wan, H.; Amoroso, A.; Joris, B.; Bouhss, A.; Calvet-Vitale*, S.; Gravier-Pelletier, C. “A sub-micromolar MraYAA Inhibitor with an aminoribosyl uridine structure and a (S,S)-tartaric diamide: Synthesis, biological evaluation and molecular modeling.” Molecules 2022, 27, 1769. https://doi.org/10.3390/molecules27061769
- Oliver, M.; Le Corre, L.; Poinsot, M.; Corio, A.; Madegard, L.; Bosco, M.; Amoroso, A.; Joris, B.; Auger, R.; Touzé, T.; Bouhss, A.; Calvet-Vitale, S.; Gravier-Pelletier, C.“Synthesis, biological evaluation and molecular modeling of urea-containing MraY inhibitors.” Org. Biomol. Chem. 2021, 19(26), 5844-5866, DOI: 10.1039/D1OB00710F.
- T. Zhang, L. Le Corre, O. Reinaud, B. Colasson “A Promising Approach for Controlling the 2nd Coordination Sphere of Biomimetic Metal Complexes: Encapsulation in a Dynamic Hydrogen-Bonded Capsule.” , Chem. Eur. J. 2021, 27, 434-443. DOI:/10.1002/chem.202004370
- Ashraf, U.; Tengo, L.; Le Corre, L.; Fournier, G.; Busca, P.; McCarthy, A. A.; Rameix-Welti, M.-A.; Gravier-Pelletier, C.; Ruigrok, R. W. H.; Jacob, Y.; et al. Destabilization of the Human RED-SMU1 Splicing Complex as a Basis for Host-Directed Antiinfluenza Strategy.” Natl. Acad. Sci. U. S. A. 2019,116(22), 10968–10977. https://doi.org/10.1073/pnas.1901214116.
- Morlot, C.; Straume, D.; Peters, K.; Hegnar, O. A.; Simon, N.;Villard, A. M.; Contreras-Martel, C.; Leisico, F.; Breukink, E.; Gravier-Pelletier, C.; Le Corre, L.; Vollmer, W.; Pietrancosta, N.; Havarstein, L. S.; Zapun. A Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae. Nat. Commun 2018, 9.
- Fer, M. J.; Corre, L. L.; Pietrancosta, N.; Evrard-Todeschi, N.;Olatunji, S.; Bouhss, A.; Calvet-Vitale, S.; Gravier-Pelletier, C. Bacterial Transferase MraY, a Source of Inspiration towards New Antibiotics. Curr Med Chem 2018, 25, 6013-6029
