Nuclear Magnetic Resonance Spectroscopy

Team Composition
Permanent Staff
M. Baudin, IR
G. Bertho, IR
C. Caradeuc, AI
F. Kateb, MCU
Students
M. El Hayek, PhD
L. Gutierrez, PhD
A. Roux, PhD
M. Soussi-Therond, PhD
C. Aldoma, M2
H. Passoubady, BUT
Post-docs
Former members

From left to right: Mosbah El Hayek, Gildas Bertho, Fatiha Kateb, Mathieu Baudin, Arthur Roux, Cédric Caradeuc, Nicolas Giraud, Léa Gutierrez.
Team Description
 The research group on NMR of Biological Substances is part of the team Bio-Spectroscopies. We develop methods that combine advanced NMR-based approaches with other physical chemical techniques including simulation tools to study complex molecular and supramolecular systems in the field of Health Sciences, targeting their dynamic and structural features. Over the last decade, we have contributed to exploring fundamental research issues at the frontier of chemistry and biology, ranging from the study of supramolecular interactions involving proteins, to the metabolic profiling of biofluids and the hyperpolarization of biological samples. These projects are carried out in the frame of several collaborations with laboratories working in the fields of organic chemistry, biochemistry, structural biology or medical research.
The research group on NMR of Biological Substances is part of the team Bio-Spectroscopies. We develop methods that combine advanced NMR-based approaches with other physical chemical techniques including simulation tools to study complex molecular and supramolecular systems in the field of Health Sciences, targeting their dynamic and structural features. Over the last decade, we have contributed to exploring fundamental research issues at the frontier of chemistry and biology, ranging from the study of supramolecular interactions involving proteins, to the metabolic profiling of biofluids and the hyperpolarization of biological samples. These projects are carried out in the frame of several collaborations with laboratories working in the fields of organic chemistry, biochemistry, structural biology or medical research.
Research Themes
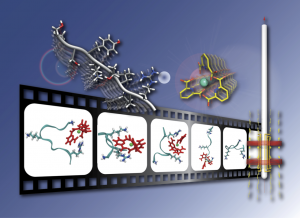
Methodological developments for ultra-high resolution NMR
 We explore original concepts that aim at enhancing resolution and sensitivity in NMR spectra of complex samples, to a point where their analysis becomes a faster, easier, and a more accurate process. We design novel pulse sequences that exceed the resolution limits of traditional NMR analyzes.
We explore original concepts that aim at enhancing resolution and sensitivity in NMR spectra of complex samples, to a point where their analysis becomes a faster, easier, and a more accurate process. We design novel pulse sequences that exceed the resolution limits of traditional NMR analyzes.
We are also working on the development of modelling and data analysis tools to make the best use of the information obtained on the biological systems under study. We demonstrate the robustness of our bioanalytical tools by applying them to study very diverse biological samples, ranging from inorganic compounds, proteins and intrinsically disordered peptides to biofluids such as urine, blood or plasma.
 Over the last years, we have carried out methodological developments to acquire Pure Shift spectra with efficient suppression of the water signal on chalenging biofluids. We have shown how the analysis of the resulting metabolic profiles allows for getting a unique insight into the metabolic pathways that are key to Lymphoma cells and for addressing the mechanisms of action of a combination of antimetabolic drugs. [1]
Over the last years, we have carried out methodological developments to acquire Pure Shift spectra with efficient suppression of the water signal on chalenging biofluids. We have shown how the analysis of the resulting metabolic profiles allows for getting a unique insight into the metabolic pathways that are key to Lymphoma cells and for addressing the mechanisms of action of a combination of antimetabolic drugs. [1]
We have also reported a new analytical workflow based on the use of a library of reference Pure Shift spectra to fit the fingerprint of each metabolite of interest and determine its concentration in biofluids. [2] We have demonstrated that this approach is robust enough to address a broad range of biological samples that are key to biomedical analyses. [3]
[1] J Proteome Res 21 (4), 1041-1051, 2022.
[2] Anal. Chem. 94, 43, 14974–14984, 2022
[3] Anal. Chem. 97, 7, 3945–3954, 2025
Dynamic and structural study of biomolecular interactions
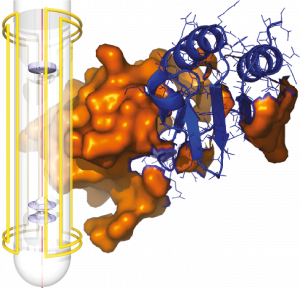
 Describing and understanding supramolecular interactions in large biological or bio-mimetic systems has become a major stake over the recent years. However, despite the considerable progress that have been achieved in this field, probing at an atomic scale protein-protein, protein-nucleic acid, protein-lipid, protein-carbohydrate interactions, or the interactions with small molecules, enzyme substrates and regulators, still constitutes a critical challenge for chemists and biochemists.
Describing and understanding supramolecular interactions in large biological or bio-mimetic systems has become a major stake over the recent years. However, despite the considerable progress that have been achieved in this field, probing at an atomic scale protein-protein, protein-nucleic acid, protein-lipid, protein-carbohydrate interactions, or the interactions with small molecules, enzyme substrates and regulators, still constitutes a critical challenge for chemists and biochemists.
We are carrying out several collaborations with the groups of Dr. Olivier Maury (Ecole Normale Supérieure de Lyon), Pr. Elise Dumont (Université Côte d’Azur, Nice) and Dr. Christelle Hureau (Laboratoire de Chimie de Coordination, Toulouse) to understand the supramolecular properties of molecules that can be used as crystallization agents of proteins, as modulators of the aggregation process of amyloid peptides, or as non-covalent paramagnetic tags to probe the structure of proteins and peptides by NMR, both in solution and in the solid state.
We notably develop an original approach combining paramagnetic NMR and molecular dynamics to probe the dynamic interaction processes between biomolecules and lanthanide complexes. [1] We have recently shown that this hybrid approach makes it possible to capture the detail of these interactions on time scales which are complementary, and to propose a global vision of the dynamic process which reconciles the points of view of the two techniques.[2]
[1] Physical Chemistry Chemical Physics, 23, 11224-11232 (2021) DOI: 10.1039/D0CP06570F
[2] Physical Chemistry Chemical Physics, 26, 14573-14581 (2024) DOI: 10.1039/D4CP00463A
Metabolomics in the field of Health Sciences
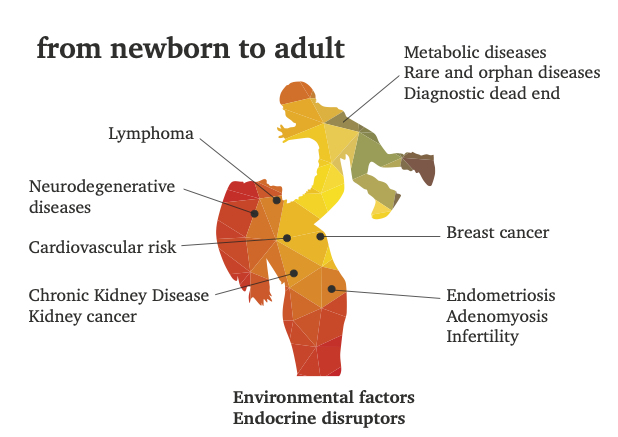 We provide medical researchers with a better understanding, based on metabolomics by NMR, of the complex interplay between the different factors that contribute to the development of diseases. Recent progress in metabolomics has improved our understanding of the system-levels effect of metabolites in biofluids, cells and tissues, using either Mass Spectrometry and/or NMR as complementary detection techniques.
We provide medical researchers with a better understanding, based on metabolomics by NMR, of the complex interplay between the different factors that contribute to the development of diseases. Recent progress in metabolomics has improved our understanding of the system-levels effect of metabolites in biofluids, cells and tissues, using either Mass Spectrometry and/or NMR as complementary detection techniques.
NMR is a highly versatile tool for obtaining metabolic profiles. It allows for analysing all the small metabolites simultaneously, with a high reproducibility. This technique is also non-destructive, robust and quantitative. Over the last decade, we have developed an expertise in deciphering the complex metabolic pathways that are involved in the evolution of diseases at every step of biological and medical research projects, from preliminary in vitro studies to clinical trials. [1]
Over the last decade, we have developed an expertise in deciphering the complex metabolic pathways that are involved in the evolution of diseases at every step of biological and medical research projects, from preliminary in vitro studies to clinical trials. [1]
Our goal is notably to bring another vision that is complementary to transcriptomics or genomics and can contribute to establish pertinent biomarkers that will help to develop new diagnosis and prognosis approaches. We can help medical researchers to better understand the mechanism of action of new therapeutic approaches, by exploring how the metabolism is correlated to the response of patients to a given treatment. In this context, our group is also developing new analytical tools for boosting resolution and sensitivity of NMR analyses and thus paving the way for new therapeutic options. [2]
[1] NMR in Biomedicine. 2023; 36(11):e5006. DOI:10.1002/nbm.5006
[2] Magn Reson Chem 2023, 1. DOI: 10.1002/mrc.5356
Dissolution Dynamic Nuclear Polarization (D-DNP) of bio-samples.
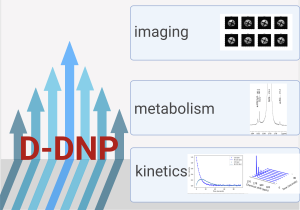 Our research activity on hyperpolarization, using dissolution dynamic nuclear polarization (D-DNP), is in line with the study of biological samples, which is a strong theme in our team. By exploiting the high signal enhancements of several orders of magnitude allowed by DNP, we seek to push the limits in terms of temporal resolution for the study of cellular processes in real time, but also to analyse complex biological samples containing low concentrations of metabolites.
Our research activity on hyperpolarization, using dissolution dynamic nuclear polarization (D-DNP), is in line with the study of biological samples, which is a strong theme in our team. By exploiting the high signal enhancements of several orders of magnitude allowed by DNP, we seek to push the limits in terms of temporal resolution for the study of cellular processes in real time, but also to analyse complex biological samples containing low concentrations of metabolites.
The starting point of our studies is the cellular metabolism of glutamine and its deregulation in the presence of certain pathologies such as cancer or cardiovascular dysfunction. [1] Initially focused on the study of enzymatic reactions in vitro and on cell extracts [2], our experiments aim at working in real time on living cells or during ex vivo MRI experiments, as well as on biological samples such as blood, urine or serum, while developing synergies with other thematics and methodological developments of the team, such as high-resolution detection sequences.
[1] Progress in Nuclear Magnetic Resonance Spectroscopy, 144-145, 15-39 (2024) DOI : 10.1016/j.pnmrs.2024.05.003
[2] ChemPhysChem 2023, 24, e202300151. DOI: 10.1002/cphc.202300151
NMR Facility at UMR 8601
Our group is hosting the NMR facility of our Research Unit, including 3 NMR spectrometers (500 MHz to 600 MHz) and a prototype polarizer for Dissolution Dynamic Nuclear Polarization (D-DNP), to improve the sensitivity of solution NMR of bio-samples.
We have recently implemented the first NMR facility in France dedicated to metabolomic analyses for Health Sciences “Metabo Paris Santé”.
Publications
2025
6. Pure Shift NMR with Solvent Suppression: a Robust and General Method for Determining Quantitative Metabolic Profiles in Biofluids.
X. Chen, C. Caradeuc, G. Bertho, C. Lucas-Torres*, N. Giraud*
Analytical Chemistry 2025, 97, 7, 3945–3954.
DOI : 10.1021/acs.analchem.4c05261
5. Lanthanide(III)-dependent hydration of the methanol dehydrogenase cofactor, pyrroloquinoline quinone.
C. Blanc, L. Oriol, T. Rajeshkumar, C. Bijani, C.L. Serpentini, N. Giraud, L. Maron, C. Hureau, E. Mathieu*
Journal of Inorganic Biochemistry, 2025, in Press
DOI : 10.1016/j.jinorgbio.2025.112924
4. Caracterization of POP mixture redistribution and identification of their molecular signature in xenografted fat mice
T. Jamay, P. Noirez, H. Djemai, L. Youssef, J. Massias, S. Ouzia, G. Cano-Sancho, P. Margaritte-Jeannin, F. Jornod, E. Blanc, X. Coumoul, Y. Guitton, B. Le Bizec, J.P. Antignac, P. Marchand, C. Lucas-Torres, N. Giraud, G. Bertho, M. Jin Kim*, K. Audouze*
Environmental Pollution, 2025, 374, 126239
DOI : 10.1016/j.envpol.2025.126239
3. Methylglyoxal-induced glycation of plasma albumin: From biomarker discovery to clinical use for prediction of new-onset diabetes in individuals with prediabetes
A. Rodrigues Oliveira, C. Chevalier, M. Wargny, V. Pakulska, C. Caradeuc, C. Cloteau, M. Letertre, N. Giraud, G. Bertho, E. Bigot-Corbel, M. Carpentier, G. Nouadje, Y. Couté, C. Le May, B. Cariou, S. Hadjadj, M. Croyal*
Clinical Chemistry 2025, in Press
DOI : 10.1093/clinchem/hvaf035
2. Metabolic Heterogeneity in Diffuse Large B-Cell Lymphoma Cells Reveals an Innovative Antimetabolic Combination Strategy
L. Lordello, S. Nuan-Aliman, K. Kielbassa-Elkadi, A. Montagne, K. Kotta, I. Martins, E. Pinto Jurado, C. Caradeuc, J. Lehmann-Che, J.A. Martinez-Liment, V. Meignin, N. Giraud, G. Kroemer, G. Bertho, C. Thieblemont, V. Baud*
Cancers 2025, 17(3), 394
DOI : 10.3390/cancers17030394
1. Identification of metabolite profiles during gout flare in a prospective gout cohort: glutamine and histidine as anti-inflammatory metabolites
C. Leroy, N. Pham, C. Caradeuc, A.L. Nguyen, F. Brial, B. Kischkeln G. Jayat, N. Giraud, F. Fenaille, L. Joosten, A. Latourte, P. Richette, F. Castelli, G. Bertho, H.K. Ea
Annals of the Rheumatic Diseases 2025, 84, 1, 111-112, Special Issue EULAR 2025: European Congress of Rheumatology
DOI: 10.1016/j.ard.2025.05.148
2024
8. Simultaneous observation of the anomerization and reaction rates of enzymatic dehydrogenation of Glucose-6-Phosphate by dissolution DNP
M. Soussi-Therond, A. Razanahoera, Y. Zhang, M. Baudin, E. Miclet, N. Giraud, D. Abergel*
Journal of the American Chemical Society 2024, 146, 50, 34651–34660.
DOI : 10.1021/jacs.4c12904
7. Les complexes de lanthanide, des sondes polyvalentes de la structure des protéines
E. Dumont, O. Maury N. Giraud*
Actualité Chimique, 2024, 499, 54
6. Revisiting the nature and pharmacodynamics of tacrolimus metabolites
R. Mevizoud, H. Aouad, F.L. Sauvage, H. Arnion, E. Pinault, J.S. Bernard, G. Bertho, N. Giraud, R. Alves de Sousa, A. Lopez-Noriega, F. Di Meo, M. Camapana, P. Marquet
Pharmacological Research 2024, 209, 107438.
DOI : 10.1016/j.phrs.2024.107438
5. Phytochemical Study of the Anthelminthic Potential of Guadeloupean Plant Biodiversity
T. Cabald, C. Marie-Magdeleine, L. Philibert, C. Caradeuc, G. Bertho, N. Giraud, G. Cebrián-Torrejón, M. Sylvestre
Pharmaceuticals 2024, 17, 774.
DOI : 10.3390/ph17060774
4. The analysis of the skeletal muscle metabolism is crucial for designing optimal exercise paradigms in type 2 diabetes mellitus
E. Abi Akar, L. Weill, M. El Khoury, C. Caradeuc, G. Bertho, S. Boutary, C. Bezier, Z. Clerc, D. Sapaly, S. Bendris, F. Cheguillaume, N. Giraud, A. A. Eid, F. Charbonnier, O. Biondi
Molecular Medicine 30, 80 (2024)
DOI: 10.1186/s10020-024-00850-7
3. Glutamine: A key player in human metabolism as revealed by hyperpolarized magnetic resonance
Dos Santos, K., Bertho, G., Baudin, M., Giraud, N.*
Progress in Nuclear Magnetic Resonance Spectroscopy (2024)
DOI : 10.1016/j.pnmrs.2024.05.003
2. One touch is all it takes: the supramolecular interaction between ubiquitin and lanthanide complexes revisited by paramagnetic NMR and molecular dynamics
K. Dos Santos, A. Bartocci, N. Gillet, S. Denis-Quanquin, A. Roux, E. Lin, Z. Xu, R. Finizola,P. Chedozeau,X. Chen, C. Caradeuc, M. Baudin, G. Bertho, F. Riobé, O. Maury, E. Dumont* and N. Giraud* ,
Physical Chemistry Chemical Physics 26, 14573-14581 (2024)
DOI: 10.1039/D4CP00463A
Selected as a 2024 PCCP HOT ARTICLE
1. Lipoproteome dysregulation precedes conversion to psychosis in ultra-high-risk subjects
M.T. Avella, G. Bertho, O. Kébir, C. Caradeuc, M.O. Krebs, N. Giraud, B. Chaumette, A. Frajerman,
Neuroscience Applied 2024, Vol. 3, Suppl. 1, 103948
DOI: 10.1016/j.nsa.2024.103948
2023
4. Present and Future of Pure Shift NMR in Metabolomics
Chen, X., Bertho, G., Caradeuc, C., Giraud, N.* and Lucas-Torres, C.*,
Magnetic Resonance in Chemistry, 2023, 61, 12, 654-673.
Metabolomics Special Issue: Frontiers in NMR Metabolomics
DOI: 10.1002/mrc.5356
Recognized as a 2023 top viewed article* in Magnetic Resonance in Chemistry
3. A Toolbox For Glutamine Use In Dissolution Dynamic Nuclear Polarization: From Enzymatic Reaction Monitoring To The Study Of Cellular Metabolic Pathways And Imaging.
Dos Santos, K, Bertho, G., Caradeuc, C., Baud, V., Montagne, A., Abergel, D., Giraud, N.* & Baudin, M.*
ChemPhysChem (2023) DOI: 10.1002/cphc.202300151
2. Guest exchange in a biomimetic ZnII cavity-complex: kinetic control by a catalytic water, through pore selection, 2nd sphere assistance, and induced-fit processes
Nyssen, N., Giraud, N., Wouters, J., Jabin, I., Leherte, L. & Reinaud, O.*
Inorganic Chemistry Frontiers, in Press (2023) DOI : 10.1039/D3QI01271A
1. NMR metabolomics study of the chronic low-dose exposure to a cocktail of Persistent Organic Pollutants
Lucas-Torres, C., Caradeuc, C., Prieur, L., Djemai, H., Layale, Y., Noirez, P., Coumoul, X., Audouze, K., Giraud, N., & Bertho, G.*
NMR in Biomedicine, in Press (2023) DOI : 10.1002/nbm.5006
2022
5. Absolute Metabolite Quantification Using Pure Shift NMR: Towards Quantitative Metabolic Profiling of Aqueous Biological Samples
Chen, X., Caradeuc, C., Montagne, A., Baud, V., Bertho, G., Lucas-Torres, C.*, Giraud, N.*
Analytical Chemistry, 2022, 94, 43, 14974–14984
DOI: 10.1021/acs.analchem.2c02823
4. Synthesis and Characterization of Novel Conformers of (E)-2-(3-nitro-H-imidazo[1,2-a]pyridin-2-ylthio)-N’-benzylideneacetohydrazide Derivatives
Evrard, A., Coulibaly, S.*, Coulibali, S *, Kouassi, S., Achi, P.A., Giraud, N. and Bertho, G.
Magnetic Resonance in Chemistry, 2022, 60, 1157-1170
DOI: 10.1002/mrc.5308
3. Lanthanide tris-dipicolinate: a do-it-all complex?
Denis Quanquin, S., D’Aléo, A., Balogh, C., N’Dala Louika, I., Salaam, J., le Guennic, B., Thieuleux, C., Camp, C., Veyre, L., Costuas, K., Rigaut, S., Le Bozec, H., Andraud, C., Dumont, E., Gilet, N., Chapelle, C., Girard, E., Giraud, N., Pilet, G., Riobé, F., Maury, O*
Actualité Chimique, 2022, 475, 12
2. Ultrahigh-Resolution NMR with Water Signal Suppression for a Deeper Understanding of the Action of Antimetabolic Drugs on Dif-fuse Large B-Cell Lymphoma
Bertho, G., Lordello, L., Chen, X., Lucas-Torres, C., Oumezziane, I.E., Caradeuc, C., Baudin, M., Nuan-Aliman, S., Thieblemont, C., Baud, V.* and Giraud, N.*
Journal of Proteome Research, 2022, 21, 4, 1041-1051
DOI: https://doi.org/10.1021/acs.jproteome.1c00914
1. Structural Analysis of Unstable Norbixin Isomers Guided by Pure Shift Nuclear Magnetic Resonance.
Bertho, G.* Oumezziane, I.E., Caradeuc, C., Guibout, L., Balducci, C., Dinan, L., Dilda, P.J., Camelo, S., Lafont, R., and Giraud, N.*
Magnetic Resonance in Chemistry, 2022, 60, 504-514
DOI: https://doi.org/10.1002/mrc.5252
2021
10. Spin Thermometry: A Straightforward Measure of Millikelvin Deuterium Spin Temperatures Achieved by Dynamic Nuclear Polarization,
B. Aghelnejad, S. Marhabaie, M. Baudin, G. Bodenhausen, and D. Carnevale
The Journal of Physical Chemistry Letters, 2021, 11 (9), 3219-3225
DOI : 10.1021/acs.jpclett.0c00713
9. Effects of Microwave Gating on Nuclear Spin Echoes in Dynamic Nuclear Polarization.
D. Guarin, D. Carnevale, M. Baudin, P. Pelupessy, D. Abergel, and G. Bodenhausen.
J. Phys. Chem. Lett., 2021, 13, 175-182
DOI : 10.1021/acs.jpclett.1c03436
8. On the Supra-LUMO Interaction: Case Study of a Sudden Change of Electronic Structure as a Functional Emergence.
Lainé, P.*, Gosset, A., Lachmanová, S.N., Cherraben, S., Bertho, G., Forté, J., Perruchot, C., Jacquot de Rouville, H.P., Pospíšil, L., Hromadová, M., Brémond, E.
Chemistry – A European Journal, 2021, 27, 17889-17899
DOI : 10.1002/chem.202103136
7. Loss of prion protein control of glucose metabolism promotes neurodegeneration in model of prion diseases.
Arnould, H., Baudouin, V., Baudry, A., Ribeiro, L.W., Ardila-Osorio, H., Pietri, M., Caradeuc, C., Soultawi, C., Williams, D., Alvarez, M., Crozet, C., Djouadi, F., Laforge, M., Bertho, G., Kellermann, O., Launay, J.M., Schmitt-Ulms, G., Schneider, B.*
PLoS Pathogens, 2021, 17(10): e1009991.
DOI : 10.1371/journal.ppat.1009991
6. Distinction between 2’- and 3’ – phosphate isomers of a fluorescent NADPH analogue led to strong inhibition of cancer cells proliferation and migration..
Lima, M., Dilly, S., Daunay, S., Abbe, P., Solier, S., Guillaumond, F., Tubiana, S.S., Escargueil, A.E., Henriques, J.A.P., Ferrand, N., Erdelmeier, I., Boucher, J.L., Bertho, G., Agranat, I., Rocchi, S., Sabbah, M., Slama-Schwok, A.*
Antioxydants, 2021, 10 (5), 723
DOI : 10.3390/antiox10050723
5. The complex metabolism of poststerone in male rats.
Balducci, C., Dinan, L., Guibout, L., Foucault, A.S., Carbonne, C., Durand, J.D., Caradeuc, C., Bertho, G., Girault, J.P., Lafont, R.
The Journal of Steroid Biochemistry and Molecular Biology, 2021, 212, 105897
DOI : 10.1016/j.jsbmb.2021.105897
4. Adenomyosis is associated with specific proton nuclear magnetic resonance (1H-NMR) serum metabolic profiles.
Bourdon, M., Santulli, P., Kateb, F., Pocate-Cheriet, K., Batteux, F., Maignien, C., Chouzenoux, S., Bordonne, C., Marcellin, L., Bertho, G.*, Chapron, C.*
Fertility and Sterility, 2021, 116 (1), 243-254
DOI : 10.1016/j.fertnstert.2021.02.031
3. Book Chapter: Analyse Métabolomique par Résonance Magnétique Nucléaire : Application aux Maladies Porphyriques
G. Bertho
in La Révolution Biotechnologique et la médecine de demain
Ducancel F., Schneider, B.
Doin Ed., ISBN: 978-2-7040-1620-4 (2021)
2. Capturing The Dynamic Association Between A Tris-Dipicolinate Lanthanide Complex And A Decapeptide: A Combined Paramagnetic NMR And Molecular Dynamics Exploration.
Denis-Quanquin, S., Bartocci, A., Szczepaniak, F., Riobé, F., Maury, O., Dumont, E.*, Giraud, N.*
Physical Chemistry Chemical Physics., 2021, 23, 11224-11232
DOI : 10.1039/D0CP06570F
1. Solid-State versus Solution Investigation of a Luminescent Chiral BINOL-Derived Bisphosphate Single-Molecule Magnet
Mattei C.A., Montigaud, V., Gendron F., Denis-Quanquin S., Dorcet V., Giraud N., Riobé F., Argouarch G., Maury O., Le Guennic B.,* Cador O., Lalli C., Pointillart F.*
Inorg. Chem. Front., 2021, 8, 947-962
DOI : 10.1039/D0QI01192D
2020
4. Mn(I) Complex Redox Potential Tunability by Remote Lewis Acid Interaction.
Srinivasan, A. Campos, J., Giraud, N., Robert M. and Rivada-Wheelaghan O.*
Dalton Trans., 2020, 49, 16623-16626
DOI : 10.1039/D0DT02467H
3. The Follicular fluid metabolome differs according to the endometriosis phenotype.
Pocate-Cheriet, K., Santulli, P., Kateb, F., Bourdon, M., Maignien, C., Batteux, F., Chouzenoux, S., Patrat, C., Wolf, J.P., Bertho, G. & Chapron, C.
Reproductive BioMedicine Online, 2020, 41, 1023-1037
DOI : 10.1016/j.rbmo.2020.09.002
2. Endometriosis Phenotypes Are Associated With Specific Serum Metabolic Profiles Determined By Proton-Nuclear Magnetic Resonance.
Maignien, C., Santulli, P., Kateb, F., Caradeuc, C., Marcellin, L., Pocate-Cheriet, K., Bourdon, M., Chouzenoux, S., Batteux, F., Bertho, G. & Chapron, C.
Reproductive BioMedicine Online, 2020, 41, 640-652
DOI : 10.1016/j.rbmo.2020.06.019
1. Multinuclear NMR in Polypeptide Liquid Crystals: Three Fertile Decades of Methodological Developments and Analytical Challenges.
Lesot, P.*, Aroulanda, C., Berdagué, P., Meddour, A., J., Merlet, D., Farjon, J., Giraud, N., & Lafon, O.
Progress in Nuclear Magnetic Resonance Spectroscopy, 2020, 116, 85-154
DOI : 10.1016/j.pnmrs.2019.10.001
2019
1. Monitoring Conformational Changes in an Enzyme Conversion Inhibitor Using Pure Shift Exchange NMR Spectroscopy.
Aloui, G., Bouabdallah, S., Baltaze, J.P., Herbert Pucheta, J.E., Touil, S., Farjon, J. & Giraud, N.*
ChemPhysChem, 2019, 20, 1738-1746
DOI : 10.1002/cphc.201900244
2. A biomimetic strategy for the selective recognition of organophosphates in 100% water: synergies of electrostatic interactions, cavity embedment and metal coordination.
Collin, S., Giraud, N., Dumont, E., & Reinaud, O.*
Organic Chemistry Frontiers, 2019, 6, 1627-1636
DOI : 10.1039/c9qo00263d
2018
1. Combining pure shift and J‐edited spectroscopies: A strategy for extracting chemical shifts and scalar couplings from highly crowded proton spectra of oligomeric saccharides.
Pitoux, D., Hu, Z., Plainchont, B., Merlet, D., Farjon, J., Bonnaffé, D. & Giraud, N.*
Magnetic Resonance in Chemistry, 2018, 56, 954-962
DOI : 10.1002/mrc.4715
2. Highly Accurate Quantitative Analysis Of Enantiomeric Mixtures From Spatially Frequency Encoded 1H NMR Spectra.
Plainchont, B., Pitoux, D., Cyrille, M. & Giraud, N.*
Analytical Chemistry, 2018, 90, 1595-1600
DOI : 10.1021/acs.analchem.7b02411
3. 1H NMR analyses of enantiomeric mixtures using chiral liquid crystals.
Farjon, J. & Giraud, N.*
Current Opinion in Colloid and Interface Science, 2018, 33, 1-8
DOI : 10.1016/j.cocis.2017.11.001
4. Real-time and non-invasive monitoring of renal endoplasmic reticulum stress in individuals with a hemodynamic impairment.
B. Fohlen, Q. Tavernier, T. Huynh, C. Caradeuc, D. Le Corre, G. Bertho, B. Cholley, N. Pallet
EBioMedicine, 2018, 27, 284-292
DOI : 10.1016/j.ebiom.2017.12.023
5. Urinary metabolic profiling of asymptomatic Acute Intermittent Porphyria using a rule-mining-based algorithm.
M. Luck, C. Schmitt, N. Talbi, L. Gouya, C. Caradeuc, H. Puy, G. Bertho*, N. Pallet*
Metabolomics, 2018, 14, 10
DOI : 10.1007/s11306-017-1305-9
6. Bacterial Transferase MraY, a Source of Inspiration towards New Antibiotics
Mickaël J. Fer, Laurent Le Corre, Nicolas Pietrancosta, Nathalie Evrard-Todeschi, Samir Olatunji, Ahmed Bouhss, Sandrine Calvet-Vitale*, Christine Gravier-Pelletier*.
Current Medicinal Chemistry, 2018, 25 , Issue 42
DOI : 10.2174/0929867325666180330095154
Till 2017
M Sarkis, MA Miteva, M Chiara Dasso Lang, M Jaouen, M-A Sari, M-O Galcéra, M Ethève-Quelquejeu, C Garbay, G Bertho*, E Braud* Insights into the interaction of high potency inhibitor IRC-083864 with phosphatase CDC25 : Binding model of CDC25 inhibitor IRC-083864 Proteins, 2017, Jan 5 doi: 101002/prot25236
M Melikian, B Eluard, G Bertho, V Baud, N Evrard-Todeschi Model of the Interaction between the NF-κB Inhibitory protein p100 and the E3 ubiquitin ligase β-TrCP based on NMR and Docking Experiments J Chem Inf Model, 2017 Jan 13 doi: 101021/acsjcim5b00409
Yurenko YP, Bazzi S, Marek R, Kozelka J Anion-π Interactions in Flavoproteins Involve a Substantial Charge-Transfer Component Chemistry 2017, 23 3246-3250
Kozelka J Lone pair-π interactions in biological systems: occurrence, function, and physical origin, Eur Biophys J 2017 May 2 doi: 101007/s00249-017-1210-1
R Balzan, L Fernandes, L Pidial, A Comment, B Tavitian, PR Vasos Pyruvate cellular uptake and enzymatic conversion probed by dissolution DNP‐NMR: the impact of overexpressed membrane transporters Magn Reson Chem, 2017, 55:579-583
2016
R Balzan, L Fernandes, A Comment, L Pidial, B Tavitian, PR Vasos Dissolution Dynamic Nuclear Polarization Instrumentation for Real-time Enzymatic Reaction Rate Measurements by NMR JoVE, 2016, e53548-e53548
M Luck, G Bertho, M Bateson, A Karras, A Yartseva, E Thervet, C Damon, N Pallet* Rule-Mining for the Early Prediction of Chronic Kidney Disease Based on Metabolomics and Multi-Source Data PLoS One 2016 Nov 18;11(11):e0166905
J-W Chiao, M Melikian, L Han, A Tsao, S K Mencher, J Fallon, G Bertho* & L G Wang* Interaction of a small molecule and STAT3-SH2 domain to block Y705 phosphorylation and inhibit lupus nephritis Biochem Pharmacol, 2016, 99, 123-131
Novotný J, Bazzi S, Marek R, Kozelka J Lone-pair-π interactions: analysis of the physical origin and biological implications Phys Chem Chem Phys 2016 Jul 28;18(28):19472-81
2015
I Anosova, S Melnik, K Tripsianes, F Kateb, I Grummt, M Sattler A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes Nucleic Acids Res 2015, 43, 5208-5220
Badri Z, Foroutan-Nejad C, Kozelka J, Marek R On the non-classical contribution in lone-pair-π interaction: IQA perspective Phys Chem Chem Phys 2015 Oct 21;17(39):26183-9
M Luck*, A Yartseva, G Bertho, E Thervet, P Beaune, N Pallet, C Damon Metabolic profiling of 1H NMR spectra in Chronic Kidney Disease with local predictive modeling Proc Int Conf Mach Learn Appl, 2015, 176-181
L Ducassou, F André, B Ramassamy, Y Xu-Li, M-A Loriot, P Beaune, G Bertho, M Lombard, D Mansuy, J-L Boucher Expression in Yeast and Discovery of New Substrates of Human Orphan Cytochrome P450 2U1: Interpretation of their Hydroxylation Regioselectivity from Docking Studies on a Protein 3D Model Biochim Biophys Acta 2015 ; 1850(7), 1426-37
2014
N Pallet, E Thervet, P Beaune, A Karras, G Bertho* The urinary metabolome of chronic kidney disease Kidney Int, 2014, 85, 1239–1240
M Carichon, N Pallet, C Schmitt, T Lefebvre, L Gouya, N Talbi, J-C Deybach, P Beaune, P Vasos, H Puy, G Bertho* The urinary metabolic fingerprint of Acute Intermittent Porphyria analyzed by 1H-NMR spectroscopy Anal Chem, 2014, 86, 2166−2174
S Téletchéa, V Stresing, S Hervouet, M Baud’huin, M-F Heymann, G Bertho, C Charrier, K Ando, D Heymann Novel RANK antagonists for the treatment of bone resorptive disease: Theoretical predictions and experimental validation J Bone Miner Res 2014, 29, 1466-1477
A Sadet, L Fernandes, F Kateb, R Balzan, PR Vasos Long-lived coherences: Improved dispersion in the frequency domain using continuous-wave and reduced-power windowed sustaining irradiation J Chem Phys 2014, 141 (5), 054203
2013
F Kateb, H Perrin, K Tripsianes, P Zou, R Spadaccini, M Bottomley, TM Franzmann, J Buchner,S Ansieau, M Sattler Structural and Functional Analysis of the DEAF-1 and BS69 MYND Domains PLoS One 2013, 8: e54715
L Fernandes, C Guerniou, I Marín‐Montesinos, M Pons, F Kateb, PR Vasos Long‐lived states in an intrinsically disordered protein domain Magn Reson Chem 2013, 51 (11), 729-733
C Decroos, V Balland, J-L Boucher, G Bertho, Y Xu, D Mansuy Towards Stable Electron Paramagnetic Resonance Oximetry Probes Synthesis, Characterization and Metabolic Evaluation of New Ester Derivatives of a Tris-(para-carboxyltetrathiaaryl)methyl (TAM) RadicalChem Res Toxicol, 2013, 26, 1561–1569
P Dansette, D Levent, A Hessani, G Bertho, D Mansuy Thiolactone Sulfoxides as New Reactive Metabolites Acting as Bis-Electrophiles: Implication in Clopidogrel and Prasugrel Bioactivation Chem Res Toxicol, 2013, 26, :794-802
C Charrier, G Bertho, O Petigny, P Moneton, R Azerad A new derivative detected in accelerated ageing of artesunate-amodiaquine fixed dose combination tablets J Pharm Biomed Anal 2013, 81-82:20-26
M Hamel, M Lecinq, M Gulea, J Kozelka Ortho-(methylsulfanyl) phenylphosphonates and derivatives: Synthesis and applications as mono-or bidentate ligands for the preparation of platinum complexes J Org Chem, 2013, Vol 745–746, 206–213
Bergès J, Fourré I, Pilmé J, Kozelka J Quantum chemical topology study of the water-platinum(II) interaction Inorg Chem 2013 Feb 4;52(3):1217-27
2012
T Suchánková, K Kubíček, J Kašpárková, V Brabec, J Kozelka Platinum–DNA interstrand crosslinks: Molecular determinants of bending and unwinding of the double helix J Inorg Biochem 2012 Mar;108:69-79
Monnet J, Kozelka J Cisplatin GG-crosslinks within single-stranded DNA: origin of the preference for left-handed helicity J Inorg Biochem 2012 Oct;115:106-12
A Mantsyzov, G Bouvier, N Evrard-Todeschi, G Bertho* Contact-based ligand-clustering approach for the identification of active compounds in virtual screening Adv Appl Bioinform Chem, 2012, doi:102147/AABCS30881
Fougeray, I Mami, G Bertho, C Legendre, P Beaune, E Thervet, N Pallet Tryptophan depletion and the kinase GCN2 mediate interferon γ-induced autophagy J Immunol, 2012, 189, 2954-2964
P Rada, A I Rojo, N Evrard-Todeschi, N G Innamorato, A Cotte, T Jaworski, J C Tobón-Velasco, H Devijver, M Flor García-Mayoral, F Van Leuven, J D Hayes, G Bertho*, A Cuadrado* Structural and functional characterization of Nrf2 degradation by the GSK-3/β-TrCP axis Mol Cell Biol, 2012, 32, 3486-3499
P Dansette, J Rosi, J Debernardi, G Bertho, D Mansuy Metabolic Activation of Prasugrel : Nature of the two Competitive Pathways Resulting in the Opening of its Thiophene RingChem Res Toxicol, 2012, 25, 1058–1065
C Chopard, T Prangé, G Bertho Naphthalene-dioxygenase catalysed cis-dihydroxylation of bicyclic azaarenes RSC Adv, 2012, 2, 605-615
PM Dansette, J Rosi, G Bertho, D Mansuy Cytochromes P450 Catalyze Both Steps of the Major Pathway of Clopidogrel Bioactivation Whereas Paraoxonase Catalyzes the Formation of a Minor Thiol Metabolite Isomer Chem Res Toxicol, 2012, 25, 348-356
2011
Kozelka, J* Evaluation of dissociation constants from competition binding experiments based on the relative binding ratio Anal Biochem, 2011,409, 66‐73
Kumpun, S ; Maria, A ; Crouzet, S ; Evrard‐Todeschi, N ; Girault, JP ; Lafont, R* Ecdysteroids from Chenopodium quinoa Willd, an ancient Andean crop of high nutritional value Food Chem, 2011, 125, 1226‐123
Kumpun, S ; Girault, JP ; Dinan, L ; Blais, C ; Maria, A ; Dauphin‐Villemant, C ; Yingyongnarongkul, B ; Suksamrarn, A ; Lafont, R* The metabolism of 20‐hydroxyecdysone in mice: relevance to pharmacological effects and gene switch applications of ecdysteroids. J Steroid Biochem Mol Biol, 2011, 126, 1‐9
Pons, J ; Tanchou, V ; Girault, JP ; Bertho, G ; Evrard‐Todeschi, N* NMR applications for identifying β‐TrCP protein‐ligand interactions Mini Rev Med Chem, 2011, 11, 283‐297
Sarkar, R ; Ahuja, P *; Vasos, PR ; Bornet, A ; Wagnieres, O ; Bodenhausen, G Long‐lived coherences for line‐narrowing in high‐field NMR Progr Nucl Magn Res Spectrosc, 2011, 59, 83‐9
Segawa, TF *; Bornet, A ; Salvi, N ; Mieville, P ; Vitzthum, V ; Carnevale, D ; Jannin, S ; Caporini, MA ; Ulzega, S ; Vasos, PR ; Rey, M ; Bodenhausen, G* Extending Timescales and Narrowing Linewidths in NMR Chimia, 2011, 65, 652‐655
Bornet, A ; Ahuja, P ; Sarkar, R ; Fernandes, L ; Hadji, S ; Lee, SY ; Haririnia, A ; Fushman, D ; Bodenhausen, G ; Vasos, PR* Long‐lived states to monitor protein unfolding by proton NMR Chem. Phys. Chem., 2011, 12, 2729‐2734
2010
Rizzato, S ; Bergès, J ; Mason, SA ; Albinati, A ; Kozelka, J* Dispersion‐driven hydrogen bonding: predicted hydrogen bond between water and platinum(II) identified by neutron diffraction Angew Chem Int Ed, 2010,49, 7440‐7443
Ahuja, P ; Sarkar, R ; Jannin, S ; Vasos, PR *; Bodenhausen, G Proton hyperpolarisation preserved in long‐lived states Chem Commun, 2010, 46, 8192‐8194
Mamadalieva, NZ ; Janibekov, AA ; Girault, JP ; Lafont, R* Two minor phytoecdysteroids of the plant Silene viridiflora. Nat Prod Commun, 2010, 5, 1579‐1582
Bouvier, G ; Evrard‐Todeschi, N ; Girault, JP ; Bertho, G* Automatic clustering of docking poses in virtual screening process using self‐organising map Bioinformatics, 2010, 26, 53‐60
2009
Kozelka, J* Molecular origin of the sequence‐dependent kinetics of reactions between cisplatin derivatives and DNA Inorg Chim Acta, 2009,362, 651–668
Téletchéa, S ; Skauge, T ; Sletten, E ; Kozelka, J* Cisplatin Adducts on a GGG Sequence within a DNA Duplex Studied by NMR Spectroscopy and Molecular Dynamics Simulations Chem. Eur. J., 2009,15, 12320 – 12337
Crouzet, S ; Maria, A ; Dinan, L ; Lafont, R ; Girault, JP* Ecdysteroids from Cyanotis longifolia Benth. (Commelinaceae). Arch Insect Biochem Physiol, 2009, 72, 194‐209
Zibareva, L ; Yeriomina, VI ; Munkhjargal, N ; Girault, JP ; Dinan, L ; Lafont, R. * The phytoecdysteroid profiles of 7 species of Silene (Caryophyllaceae). Arch Insect Biochem Physiol, 2009, 72, 234‐248
2008
Ho, R ; Girault, JP ; Cousteau, PY ; Bianchini, JP ; Raharivelomanana, P ; Lafont, R* Isolation of a new class of ecdysteroid conjugates (glucosyl‐ferulates) using a combination of liquid chromatographic methods. J Chromatogr Sci, 2008, 46, 102‐110
Bertho, G *;. Bouvier, G ; Hui Bon Hoa, G ; Girault, JP* The key‐role of tyrosine 155 in the mechanism of prion transconformation as highlighted by a study of sheep mutant peptides. Peptides, 2008, 29, 1073‐1084
Pons, J ; Evrard‐Todeschi, N ; Bertho, G ; Gharbi‐Benarous, J ; Tanchou, V ; Benarous, R ; Girault, JP* Transfer‐NMR and Docking Studies Identify the Binding of the Peptide Derived from Activating Transcription Factor 4 to Protein Ubiquitin Ligase beta‐TrCP. Competition STD‐NMR with beta‐Catenin Biochemistry, 2008, 47, 14‐29 (Hot article)
Evrard‐Todeschi, N ; Pons, J ; Gharbi‐Benarous, J ; Bertho, G ; Benarous, R ; Girault, JP* Structure of the Complex between Phosphorylated Substrates and the beta‐TrCP Ubiquitine Ligase Receptor: A combined NMR, Molecular Modelling and Docking Approach J Chem Inf Model, 2008, 48, 2350‐2361
2007
Pons, J ; Evrard‐Todeschi, N ; Bertho, G ; Gharbi‐Benarous, J ; Sonois, V ; Benarous, R ; Girault, JP* Structural studies on 24P‐IkBa peptide derived from a human IkB‐a protein‐related with the inhibition of the transcription factor nuclear NFkB activity Biochemistry, 2007, 46, 2958‐2972
Pons, J ; Evrard‐Todeschi, N ; Bertho, G ; Gharbi‐Benarous, J ; Benarous, R ; Girault, JP* Phosphorylation‐Dependent Structure of ATF4 Peptides derived from a Human ATF4 Protein, a Member of the Family of Transcription Factors Peptides, 2007, 28, 2253‐2267
Snogan, E ; Vahirua‐Lechat, I ; Ho, R ; Bertho, G ; Girault, JP ; Ortiga, S ; Maria, A ; Lafont R* Ecdysteroids from the Medicinal Fern Microsorum scolopendria (Burm. f.) Phytochem Anal., 2007, 18, 441‐450
Aitken, DJ ; Albinati, A ; Gutier, A ; Husson, H‐P ; Morgant, G ; Nguyen‐Huy, D ; Kozelka, J *; Lemoine, P ; Ongeri, S ; Rizzato, S ; Viossat, B Platinum(II) and Palladium(II) Complexes with N‐Aminoguanidine Eur J Inorg Chem, 2007, 3327 ‐ 3334
Hamel, M ; Rizzato, S ; Lecinq, M ; Sene, A ; Vazeux, M ; Gulea, M ; Albinati, A ; Kozelka, J* Study of Intramolecular Competition between Carboxylate and Phosphonate for PtII with the Aid of a Novel Tridentate Carboxylato‐Thioether‐Phosphonato Ligand Chem. Eur. J., 2007,13, 5441‐5449
Nuclear Magnetic Resonance Spectroscopy
Team composition

Team Description
 The research group on NMR of Biological Substances is part of the team Bio-Spectroscopies. We develop methods that combine advanced NMR-based approaches with other physical chemical techniques including simulation tools to study complex molecular and supramolecular systems in the field of Health Sciences, targeting their dynamic and structural features. Over the last decade, we have contributed to exploring fundamental research issues at the frontier of chemistry and biology, ranging from the study of supramolecular interactions involving proteins, to the metabolic profiling of biofluids and the hyperpolarization of biological samples. These projects are carried out in the frame of several collaborations with laboratories working in the fields of organic chemistry, biochemistry, structural biology or medical research.
The research group on NMR of Biological Substances is part of the team Bio-Spectroscopies. We develop methods that combine advanced NMR-based approaches with other physical chemical techniques including simulation tools to study complex molecular and supramolecular systems in the field of Health Sciences, targeting their dynamic and structural features. Over the last decade, we have contributed to exploring fundamental research issues at the frontier of chemistry and biology, ranging from the study of supramolecular interactions involving proteins, to the metabolic profiling of biofluids and the hyperpolarization of biological samples. These projects are carried out in the frame of several collaborations with laboratories working in the fields of organic chemistry, biochemistry, structural biology or medical research.
Research Themes
Methodological developments for ultra-high resolution NMR
 We explore original concepts that aim at enhancing resolution and sensitivity in NMR spectra of complex samples, to a point where their analysis becomes a faster, easier, and a more accurate process. We design novel pulse sequences that exceed the resolution limits of traditional NMR analyzes.
We explore original concepts that aim at enhancing resolution and sensitivity in NMR spectra of complex samples, to a point where their analysis becomes a faster, easier, and a more accurate process. We design novel pulse sequences that exceed the resolution limits of traditional NMR analyzes.
We are also working on the development of modelling and data analysis tools to make the best use of the information obtained on the biological systems under study. We demonstrate the robustness of our bioanalytical tools by applying them to study very diverse biological samples, ranging from inorganic compounds, proteins and intrinsically disordered peptides to biofluids such as urine, blood or plasma.
 Over the last years, we have carried out methodological developments to acquire Pure Shift spectra with efficient suppression of the water signal on chalenging biofluids. We have shown how the analysis of the resulting metabolic profiles allows for getting a unique insight into the metabolic pathways that are key to Lymphoma cells and for addressing the mechanisms of action of a combination of antimetabolic drugs. [1]
Over the last years, we have carried out methodological developments to acquire Pure Shift spectra with efficient suppression of the water signal on chalenging biofluids. We have shown how the analysis of the resulting metabolic profiles allows for getting a unique insight into the metabolic pathways that are key to Lymphoma cells and for addressing the mechanisms of action of a combination of antimetabolic drugs. [1]
We have also reported a new analytical workflow based on the use of a library of reference Pure Shift spectra to fit the fingerprint of each metabolite of interest and determine its concentration in biofluids. [2] We have demonstrated that this approach is robust enough to address a broad range of biological samples that are key to biomedical analyses. [3]
[1] J Proteome Res 21 (4), 1041-1051, 2022.
[2] Anal. Chem. 94, 43, 14974–14984, 2022
[3] Anal. Chem. 97, 7, 3945–3954, 2025
Dynamic and structural study of biomolecular interactions
 Describing and understanding supramolecular interactions in large biological or bio-mimetic systems has become a major stake over the recent years. However, despite the considerable progress that have been achieved in this field, probing at an atomic scale protein-protein, protein-nucleic acid, protein-lipid, protein-carbohydrate interactions, or the interactions with small molecules, enzyme substrates and regulators, still constitutes a critical challenge for chemists and biochemists.
Describing and understanding supramolecular interactions in large biological or bio-mimetic systems has become a major stake over the recent years. However, despite the considerable progress that have been achieved in this field, probing at an atomic scale protein-protein, protein-nucleic acid, protein-lipid, protein-carbohydrate interactions, or the interactions with small molecules, enzyme substrates and regulators, still constitutes a critical challenge for chemists and biochemists.
We are carrying out several collaborations with the groups of Dr. Olivier Maury (Ecole Normale Supérieure de Lyon), Pr. Elise Dumont (Université Côte d’Azur, Nice) and Dr. Christelle Hureau (Laboratoire de Chimie de Coordination, Toulouse) to understand the supramolecular properties of molecules that can be used as crystallization agents of proteins, as modulators of the aggregation process of amyloid peptides, or as non-covalent paramagnetic tags to probe the structure of proteins and peptides by NMR, both in solution and in the solid state.
We notably develop an original approach combining paramagnetic NMR and molecular dynamics to probe the dynamic interaction processes between biomolecules and lanthanide complexes. [1] We have recently shown that this hybrid approach makes it possible to capture the detail of these interactions on time scales which are complementary, and to propose a global vision of the dynamic process which reconciles the points of view of the two techniques. [2]
[1] Physical Chemistry Chemical Physics, 23, 11224-11232 (2021) DOI: 10.1039/D0CP06570F
[2] Physical Chemistry Chemical Physics, 26, 14573-14581 (2024) DOI: 10.1039/D4CP00463A
Metabolomics in the field of Health Sciences
 We provide medical researchers with a better understanding, based on metabolomics by NMR, of the complex interplay between the different factors that contribute to the development of diseases. Recent progress in metabolomics has improved our understanding of the system-levels effect of metabolites in biofluids, cells and tissues, using either Mass Spectrometry and/or NMR as complementary detection techniques.
We provide medical researchers with a better understanding, based on metabolomics by NMR, of the complex interplay between the different factors that contribute to the development of diseases. Recent progress in metabolomics has improved our understanding of the system-levels effect of metabolites in biofluids, cells and tissues, using either Mass Spectrometry and/or NMR as complementary detection techniques.
NMR is a highly versatile tool for obtaining metabolic profiles. It allows for analysing all the small metabolites simultaneously, with a high reproducibility. This technique is also non-destructive, robust and quantitative. Over the last decade, we have developed an expertise in deciphering the complex metabolic pathways that are involved in the evolution of diseases at every step of biological and medical research projects, from preliminary in vitro studies to clinical trials. [1]
Over the last decade, we have developed an expertise in deciphering the complex metabolic pathways that are involved in the evolution of diseases at every step of biological and medical research projects, from preliminary in vitro studies to clinical trials. [1]
Our goal is notably to bring another vision that is complementary to transcriptomics or genomics and can contribute to establish pertinent biomarkers that will help to develop new diagnosis and prognosis approaches.
We can help medical researchers to better understand the mechanism of action of new therapeutic approaches, by exploring how the metabolism is correlated to the response of patients to a given treatment. In this context, our group is also developing new analytical tools for boosting resolution and sensitivity of NMR analyses and thus paving the way for new therapeutic options. [2]
[1] NMR in Biomedicine. 2023; 36(11):e5006. DOI:10.1002/nbm.5006
[2] Magn Reson Chem 2023, 1. DOI: 10.1002/mrc.5356
Dissolution Dynamic Nuclear Polarization (D-DNP) of bio-samples.
 Our research activity on hyperpolarization, using dissolution dynamic nuclear polarization (D-DNP), is in line with the study of biological samples, which is a strong theme in our team. By exploiting the high signal enhancements of several orders of magnitude allowed by DNP, we seek to push the limits in terms of temporal resolution for the study of cellular processes in real time, but also to analyse complex biological samples containing low concentrations of metabolites.
Our research activity on hyperpolarization, using dissolution dynamic nuclear polarization (D-DNP), is in line with the study of biological samples, which is a strong theme in our team. By exploiting the high signal enhancements of several orders of magnitude allowed by DNP, we seek to push the limits in terms of temporal resolution for the study of cellular processes in real time, but also to analyse complex biological samples containing low concentrations of metabolites.
The starting point of our studies is the cellular metabolism of glutamine [1] and its deregulation in the presence of certain pathologies such as cancer or cardiovascular dysfunction. [2] Initially focused on the study of enzymatic reactions in vitro and on cell extracts, our experiments aim at working in real time on living cells or during ex vivo MRI experiments, as well as on biological samples such as blood, urine or serum, while developing synergies with other thematics and methodological developments of the team, such as high-resolution detection sequences.
[1] Progress in Nuclear Magnetic Resonance Spectroscopy, 144-145, 15-39 (2024) DOI : 10.1016/j.pnmrs.2024.05.003
[2] ChemPhysChem 2023, 24, e202300151. DOI: 10.1002/cphc.202300151
NMR Facility at UMR 8601
Our group is hosting the NMR facility of our Research Unit, including 3 NMR spectrometers (500 MHz to 600 MHz) and a prototype polarizer for Dissolution Dynamic Nuclear Polarization (D-DNP), to improve the sensitivity of solution NMR of bio-samples.

We have recently implemented the first NMR facility in France dedicated to metabolomic analyses for Health Sciences “Metabo Paris Santé”.
2025
6. Pure Shift NMR with Solvent Suppression: a Robust and General Method for Determining Quantitative Metabolic Profiles in Biofluids.
X. Chen, C. Caradeuc, G. Bertho, C. Lucas-Torres*, N. Giraud*
Analytical Chemistry 2025, 97, 7, 3945–3954.
DOI : 10.1021/acs.analchem.4c05261
5. Lanthanide(III)-dependent hydration of the methanol dehydrogenase cofactor, pyrroloquinoline quinone.
C. Blanc, L. Oriol, T. Rajeshkumar, C. Bijani, C.L. Serpentini, N. Giraud, L. Maron, C. Hureau, E. Mathieu*
Journal of Inorganic Biochemistry, 2025, in Press
DOI : 10.1016/j.jinorgbio.2025.112924
4. Caracterization of POP mixture redistribution and identification of their molecular signature in xenografted fat mice
T. Jamay, P. Noirez, H. Djemai, L. Youssef, J. Massias, S. Ouzia, G. Cano-Sancho, P. Margaritte-Jeannin, F. Jornod, E. Blanc, X. Coumoul, Y. Guitton, B. Le Bizec, J.P. Antignac, P. Marchand, C. Lucas-Torres, N. Giraud, G. Bertho, M. Jin Kim*, K. Audouze*
Environmental Pollution, 2025, 374, 126239
DOI : 10.1016/j.envpol.2025.126239
3. Methylglyoxal-induced glycation of plasma albumin: From biomarker discovery to clinical use for prediction of new-onset diabetes in individuals with prediabetes
A. Rodrigues Oliveira, C. Chevalier, M. Wargny, V. Pakulska, C. Caradeuc, C. Cloteau, M. Letertre, N. Giraud, G. Bertho, E. Bigot-Corbel, M. Carpentier, G. Nouadje, Y. Couté, C. Le May, B. Cariou, S. Hadjadj, M. Croyal*
Clinical Chemistry 2025, in Press
DOI : 10.1093/clinchem/hvaf035
2. Metabolic Heterogeneity in Diffuse Large B-Cell Lymphoma Cells Reveals an Innovative Antimetabolic Combination Strategy
L. Lordello, S. Nuan-Aliman, K. Kielbassa-Elkadi, A. Montagne, K. Kotta, I. Martins, E. Pinto Jurado, C. Caradeuc, J. Lehmann-Che, J.A. Martinez-Liment, V. Meignin, N. Giraud, G. Kroemer, G. Bertho, C. Thieblemont, V. Baud*
Cancers 2025, 17(3), 394
DOI : 10.3390/cancers17030394
1. Identification of metabolite profiles during gout flare in a prospective gout cohort: glutamine and histidine as anti-inflammatory metabolites
C. Leroy, N. Pham, C. Caradeuc, A.L. Nguyen, F. Brial, B. Kischkeln G. Jayat, N. Giraud, F. Fenaille, L. Joosten, A. Latourte, P. Richette, F. Castelli, G. Bertho, H.K. Ea
Annals of the Rheumatic Diseases 2025, 84, 1, 111-112, Special Issue EULAR 2025: European Congress of Rheumatology
DOI: 10.1016/j.ard.2025.05.148
2024
8. Simultaneous observation of the anomerization and reaction rates of enzymatic dehydrogenation of Glucose-6-Phosphate by dissolution DNP
M. Soussi-Therond, A. Razanahoera, Y. Zhang, M. Baudin, E. Miclet, N. Giraud, D. Abergel*
Journal of the American Chemical Society 2024, 146, 50, 34651–34660.
DOI : 10.1021/jacs.4c12904
7. Les complexes de lanthanide, des sondes polyvalentes de la structure des protéines
E. Dumont, O. Maury N. Giraud*
Actualité Chimique, 2024, 499, 54
6. Revisiting the nature and pharmacodynamics of tacrolimus metabolites
R. Mevizoud, H. Aouad, F.L. Sauvage, H. Arnion, E. Pinault, J.S. Bernard, G. Bertho, N. Giraud, R. Alves de Sousa, A. Lopez-Noriega, F. Di Meo, M. Camapana, P. Marquet
Pharmacological Research 2024, 209, 107438.
DOI : 10.1016/j.phrs.2024.107438
5. Phytochemical Study of the Anthelminthic Potential of Guadeloupean Plant Biodiversity
T. Cabald, C. Marie-Magdeleine, L. Philibert, C. Caradeuc, G. Bertho, N. Giraud, G. Cebrián-Torrejón, M. Sylvestre
Pharmaceuticals 2024, 17, 774.
DOI : 10.3390/ph17060774
4. The analysis of the skeletal muscle metabolism is crucial for designing optimal exercise paradigms in type 2 diabetes mellitus
E. Abi Akar, L. Weill, M. El Khoury, C. Caradeuc, G. Bertho, S. Boutary, C. Bezier, Z. Clerc, D. Sapaly, S. Bendris, F. Cheguillaume, N. Giraud, A. A. Eid, F. Charbonnier, O. Biondi
Molecular Medicine 30, 80 (2024)
DOI: 10.1186/s10020-024-00850-7
3. Glutamine: A key player in human metabolism as revealed by hyperpolarized magnetic resonance
Dos Santos, K., Bertho, G., Baudin, M., Giraud, N.*
Progress in Nuclear Magnetic Resonance Spectroscopy (2024)
DOI : 10.1016/j.pnmrs.2024.05.003
2. One touch is all it takes: the supramolecular interaction between ubiquitin and lanthanide complexes revisited by paramagnetic NMR and molecular dynamics
K. Dos Santos, A. Bartocci, N. Gillet, S. Denis-Quanquin, A. Roux, E. Lin, Z. Xu, R. Finizola,P. Chedozeau,X. Chen, C. Caradeuc, M. Baudin, G. Bertho, F. Riobé, O. Maury, E. Dumont* and N. Giraud* ,
Physical Chemistry Chemical Physics 26, 14573-14581 (2024)
DOI: 10.1039/D4CP00463A
Selected as a 2024 PCCP HOT ARTICLE
1. Lipoproteome dysregulation precedes conversion to psychosis in ultra-high-risk subjects
M.T. Avella, G. Bertho, O. Kébir, C. Caradeuc, M.O. Krebs, N. Giraud, B. Chaumette, A. Frajerman,
Neuroscience Applied 2024, Vol. 3, Suppl. 1, 103948
DOI: 10.1016/j.nsa.2024.103948
2023
4. Present and Future of Pure Shift NMR in Metabolomics
Chen, X., Bertho, G., Caradeuc, C., Giraud, N.* and Lucas-Torres, C.*,
Magnetic Resonance in Chemistry, 2023, 61, 12, 654-673.
Metabolomics Special Issue: Frontiers in NMR Metabolomics
DOI: 10.1002/mrc.5356
Recognized as a 2023 top viewed article* in Magnetic Resonance in Chemistry
3. A Toolbox For Glutamine Use In Dissolution Dynamic Nuclear Polarization: From Enzymatic Reaction Monitoring To The Study Of Cellular Metabolic Pathways And Imaging.
Dos Santos, K, Bertho, G., Caradeuc, C., Baud, V., Montagne, A., Abergel, D., Giraud, N.* & Baudin, M.*
ChemPhysChem (2023) DOI: 10.1002/cphc.202300151
2. Guest exchange in a biomimetic ZnII cavity-complex: kinetic control by a catalytic water, through pore selection, 2nd sphere assistance, and induced-fit processes
Nyssen, N., Giraud, N., Wouters, J., Jabin, I., Leherte, L. & Reinaud, O.*
Inorganic Chemistry Frontiers, in Press (2023) DOI : 10.1039/D3QI01271A
1. NMR metabolomics study of the chronic low-dose exposure to a cocktail of Persistent Organic Pollutants
Lucas-Torres, C., Caradeuc, C., Prieur, L., Djemai, H., Layale, Y., Noirez, P., Coumoul, X., Audouze, K., Giraud, N., & Bertho, G.*
NMR in Biomedicine, in Press (2023) DOI : 10.1002/nbm.5006
2022
5. Absolute Metabolite Quantification Using Pure Shift NMR: Towards Quantitative Metabolic Profiling of Aqueous Biological Samples
Chen, X., Caradeuc, C., Montagne, A., Baud, V., Bertho, G., Lucas-Torres, C.*, Giraud, N.*
Analytical Chemistry, 2022, 94, 43, 14974–14984
DOI: 10.1021/acs.analchem.2c02823
4. Synthesis and Characterization of Novel Conformers of (E)-2-(3-nitro-H-imidazo[1,2-a]pyridin-2-ylthio)-N’-benzylideneacetohydrazide Derivatives
Evrard, A., Coulibaly, S.*, Coulibali, S *, Kouassi, S., Achi, P.A., Giraud, N. and Bertho, G.
Magnetic Resonance in Chemistry, 2022, 60, 1157-1170
DOI: 10.1002/mrc.5308
3. Lanthanide tris-dipicolinate: a do-it-all complex?
Denis Quanquin, S., D’Aléo, A., Balogh, C., N’Dala Louika, I., Salaam, J., le Guennic, B., Thieuleux, C., Camp, C., Veyre, L., Costuas, K., Rigaut, S., Le Bozec, H., Andraud, C., Dumont, E., Gilet, N., Chapelle, C., Girard, E., Giraud, N., Pilet, G., Riobé, F., Maury, O*
Actualité Chimique, 2022, 475, 12
2. Ultrahigh-Resolution NMR with Water Signal Suppression for a Deeper Understanding of the Action of Antimetabolic Drugs on Dif-fuse Large B-Cell Lymphoma
Bertho, G., Lordello, L., Chen, X., Lucas-Torres, C., Oumezziane, I.E., Caradeuc, C., Baudin, M., Nuan-Aliman, S., Thieblemont, C., Baud, V.* and Giraud, N.*
Journal of Proteome Research, 2022, 21, 4, 1041-1051
DOI: https://doi.org/10.1021/acs.jproteome.1c00914
1. Structural Analysis of Unstable Norbixin Isomers Guided by Pure Shift Nuclear Magnetic Resonance.
Bertho, G.* Oumezziane, I.E., Caradeuc, C., Guibout, L., Balducci, C., Dinan, L., Dilda, P.J., Camelo, S., Lafont, R., and Giraud, N.*
Magnetic Resonance in Chemistry, 2022, 60, 504-514
DOI: https://doi.org/10.1002/mrc.5252
2021
10. Spin Thermometry: A Straightforward Measure of Millikelvin Deuterium Spin Temperatures Achieved by Dynamic Nuclear Polarization,
B. Aghelnejad, S. Marhabaie, M. Baudin, G. Bodenhausen, and D. Carnevale
The Journal of Physical Chemistry Letters, 2021, 11 (9), 3219-3225
DOI : 10.1021/acs.jpclett.0c00713
9. Effects of Microwave Gating on Nuclear Spin Echoes in Dynamic Nuclear Polarization.
D. Guarin, D. Carnevale, M. Baudin, P. Pelupessy, D. Abergel, and G. Bodenhausen.
J. Phys. Chem. Lett., 2021, 13, 175-182
DOI : 10.1021/acs.jpclett.1c03436
8. On the Supra-LUMO Interaction: Case Study of a Sudden Change of Electronic Structure as a Functional Emergence.
Lainé, P.*, Gosset, A., Lachmanová, S.N., Cherraben, S., Bertho, G., Forté, J., Perruchot, C., Jacquot de Rouville, H.P., Pospíšil, L., Hromadová, M., Brémond, E.
Chemistry – A European Journal, 2021, 27, 17889-17899
DOI : 10.1002/chem.202103136
7. Loss of prion protein control of glucose metabolism promotes neurodegeneration in model of prion diseases.
Arnould, H., Baudouin, V., Baudry, A., Ribeiro, L.W., Ardila-Osorio, H., Pietri, M., Caradeuc, C., Soultawi, C., Williams, D., Alvarez, M., Crozet, C., Djouadi, F., Laforge, M., Bertho, G., Kellermann, O., Launay, J.M., Schmitt-Ulms, G., Schneider, B.*
PLoS Pathogens, 2021, 17(10): e1009991.
DOI : 10.1371/journal.ppat.1009991
6. Distinction between 2’- and 3’ – phosphate isomers of a fluorescent NADPH analogue led to strong inhibition of cancer cells proliferation and migration..
Lima, M., Dilly, S., Daunay, S., Abbe, P., Solier, S., Guillaumond, F., Tubiana, S.S., Escargueil, A.E., Henriques, J.A.P., Ferrand, N., Erdelmeier, I., Boucher, J.L., Bertho, G., Agranat, I., Rocchi, S., Sabbah, M., Slama-Schwok, A.*
Antioxydants, 2021, 10 (5), 723
DOI : 10.3390/antiox10050723
5. The complex metabolism of poststerone in male rats.
Balducci, C., Dinan, L., Guibout, L., Foucault, A.S., Carbonne, C., Durand, J.D., Caradeuc, C., Bertho, G., Girault, J.P., Lafont, R.
The Journal of Steroid Biochemistry and Molecular Biology, 2021, 212, 105897
DOI : 10.1016/j.jsbmb.2021.105897
4. Adenomyosis is associated with specific proton nuclear magnetic resonance (1H-NMR) serum metabolic profiles.
Bourdon, M., Santulli, P., Kateb, F., Pocate-Cheriet, K., Batteux, F., Maignien, C., Chouzenoux, S., Bordonne, C., Marcellin, L., Bertho, G.*, Chapron, C.*
Fertility and Sterility, 2021, 116 (1), 243-254
DOI : 10.1016/j.fertnstert.2021.02.031
3. Book Chapter: Analyse Métabolomique par Résonance Magnétique Nucléaire : Application aux Maladies Porphyriques
G. Bertho
in La Révolution Biotechnologique et la médecine de demain
Ducancel F., Schneider, B.
Doin Ed., ISBN: 978-2-7040-1620-4 (2021)
2. Capturing The Dynamic Association Between A Tris-Dipicolinate Lanthanide Complex And A Decapeptide: A Combined Paramagnetic NMR And Molecular Dynamics Exploration.
Denis-Quanquin, S., Bartocci, A., Szczepaniak, F., Riobé, F., Maury, O., Dumont, E.*, Giraud, N.*
Physical Chemistry Chemical Physics., 2021, 23, 11224-11232
DOI : 10.1039/D0CP06570F
1. Solid-State versus Solution Investigation of a Luminescent Chiral BINOL-Derived Bisphosphate Single-Molecule Magnet
Mattei C.A., Montigaud, V., Gendron F., Denis-Quanquin S., Dorcet V., Giraud N., Riobé F., Argouarch G., Maury O., Le Guennic B.,* Cador O., Lalli C., Pointillart F.*
Inorg. Chem. Front., 2021, 8, 947-962
DOI : 10.1039/D0QI01192D
2020
4. Mn(I) Complex Redox Potential Tunability by Remote Lewis Acid Interaction.
Srinivasan, A. Campos, J., Giraud, N., Robert M. and Rivada-Wheelaghan O.*
Dalton Trans., 2020, 49, 16623-16626
DOI : 10.1039/D0DT02467H
3. The Follicular fluid metabolome differs according to the endometriosis phenotype.
Pocate-Cheriet, K., Santulli, P., Kateb, F., Bourdon, M., Maignien, C., Batteux, F., Chouzenoux, S., Patrat, C., Wolf, J.P., Bertho, G. & Chapron, C.
Reproductive BioMedicine Online, 2020, 41, 1023-1037
DOI : 10.1016/j.rbmo.2020.09.002
2. Endometriosis Phenotypes Are Associated With Specific Serum Metabolic Profiles Determined By Proton-Nuclear Magnetic Resonance.
Maignien, C., Santulli, P., Kateb, F., Caradeuc, C., Marcellin, L., Pocate-Cheriet, K., Bourdon, M., Chouzenoux, S., Batteux, F., Bertho, G. & Chapron, C.
Reproductive BioMedicine Online, 2020, 41, 640-652
DOI : 10.1016/j.rbmo.2020.06.019
1. Multinuclear NMR in Polypeptide Liquid Crystals: Three Fertile Decades of Methodological Developments and Analytical Challenges.
Lesot, P.*, Aroulanda, C., Berdagué, P., Meddour, A., J., Merlet, D., Farjon, J., Giraud, N., & Lafon, O.
Progress in Nuclear Magnetic Resonance Spectroscopy, 2020, 116, 85-154
DOI : 10.1016/j.pnmrs.2019.10.001
2019
1. Monitoring Conformational Changes in an Enzyme Conversion Inhibitor Using Pure Shift Exchange NMR Spectroscopy.
Aloui, G., Bouabdallah, S., Baltaze, J.P., Herbert Pucheta, J.E., Touil, S., Farjon, J. & Giraud, N.*
ChemPhysChem, 2019, 20, 1738-1746
DOI : 10.1002/cphc.201900244
2. A biomimetic strategy for the selective recognition of organophosphates in 100% water: synergies of electrostatic interactions, cavity embedment and metal coordination.
Collin, S., Giraud, N., Dumont, E., & Reinaud, O.*
Organic Chemistry Frontiers, 2019, 6, 1627-1636
DOI : 10.1039/c9qo00263d
2018
1. Combining pure shift and J‐edited spectroscopies: A strategy for extracting chemical shifts and scalar couplings from highly crowded proton spectra of oligomeric saccharides.
Pitoux, D., Hu, Z., Plainchont, B., Merlet, D., Farjon, J., Bonnaffé, D. & Giraud, N.*
Magnetic Resonance in Chemistry, 2018, 56, 954-962
DOI : 10.1002/mrc.4715
2. Highly Accurate Quantitative Analysis Of Enantiomeric Mixtures From Spatially Frequency Encoded 1H NMR Spectra.
Plainchont, B., Pitoux, D., Cyrille, M. & Giraud, N.*
Analytical Chemistry, 2018, 90, 1595-1600
DOI : 10.1021/acs.analchem.7b02411
3. 1H NMR analyses of enantiomeric mixtures using chiral liquid crystals.
Farjon, J. & Giraud, N.*
Current Opinion in Colloid and Interface Science, 2018, 33, 1-8
DOI : 10.1016/j.cocis.2017.11.001
4. Real-time and non-invasive monitoring of renal endoplasmic reticulum stress in individuals with a hemodynamic impairment.
B. Fohlen, Q. Tavernier, T. Huynh, C. Caradeuc, D. Le Corre, G. Bertho, B. Cholley, N. Pallet
EBioMedicine, 2018, 27, 284-292
DOI : 10.1016/j.ebiom.2017.12.023
5. Urinary metabolic profiling of asymptomatic Acute Intermittent Porphyria using a rule-mining-based algorithm.
M. Luck, C. Schmitt, N. Talbi, L. Gouya, C. Caradeuc, H. Puy, G. Bertho*, N. Pallet*
Metabolomics, 2018, 14, 10
DOI : 10.1007/s11306-017-1305-9
6. Bacterial Transferase MraY, a Source of Inspiration towards New Antibiotics
Mickaël J. Fer, Laurent Le Corre, Nicolas Pietrancosta, Nathalie Evrard-Todeschi, Samir Olatunji, Ahmed Bouhss, Sandrine Calvet-Vitale*, Christine Gravier-Pelletier*.
Current Medicinal Chemistry, 2018, 25 , Issue 42
DOI : 10.2174/0929867325666180330095154
Till 2017
M Sarkis, MA Miteva, M Chiara Dasso Lang, M Jaouen, M-A Sari, M-O Galcéra, M Ethève-Quelquejeu, C Garbay, G Bertho*, E Braud* Insights into the interaction of high potency inhibitor IRC-083864 with phosphatase CDC25 : Binding model of CDC25 inhibitor IRC-083864 Proteins, 2017, Jan 5 doi: 101002/prot25236
M Melikian, B Eluard, G Bertho, V Baud, N Evrard-Todeschi Model of the Interaction between the NF-κB Inhibitory protein p100 and the E3 ubiquitin ligase β-TrCP based on NMR and Docking Experiments J Chem Inf Model, 2017 Jan 13 doi: 101021/acsjcim5b00409
Yurenko YP, Bazzi S, Marek R, Kozelka J Anion-π Interactions in Flavoproteins Involve a Substantial Charge-Transfer Component Chemistry 2017, 23 3246-3250
Kozelka J Lone pair-π interactions in biological systems: occurrence, function, and physical origin, Eur Biophys J 2017 May 2 doi: 101007/s00249-017-1210-1
R Balzan, L Fernandes, L Pidial, A Comment, B Tavitian, PR Vasos Pyruvate cellular uptake and enzymatic conversion probed by dissolution DNP‐NMR: the impact of overexpressed membrane transporters Magn Reson Chem, 2017, 55:579-583
2016
R Balzan, L Fernandes, A Comment, L Pidial, B Tavitian, PR Vasos Dissolution Dynamic Nuclear Polarization Instrumentation for Real-time Enzymatic Reaction Rate Measurements by NMR JoVE, 2016, e53548-e53548
M Luck, G Bertho, M Bateson, A Karras, A Yartseva, E Thervet, C Damon, N Pallet* Rule-Mining for the Early Prediction of Chronic Kidney Disease Based on Metabolomics and Multi-Source Data PLoS One 2016 Nov 18;11(11):e0166905
J-W Chiao, M Melikian, L Han, A Tsao, S K Mencher, J Fallon, G Bertho* & L G Wang* Interaction of a small molecule and STAT3-SH2 domain to block Y705 phosphorylation and inhibit lupus nephritis Biochem Pharmacol, 2016, 99, 123-131
Novotný J, Bazzi S, Marek R, Kozelka J Lone-pair-π interactions: analysis of the physical origin and biological implications Phys Chem Chem Phys 2016 Jul 28;18(28):19472-81
2015
I Anosova, S Melnik, K Tripsianes, F Kateb, I Grummt, M Sattler A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes Nucleic Acids Res 2015, 43, 5208-5220
Badri Z, Foroutan-Nejad C, Kozelka J, Marek R On the non-classical contribution in lone-pair-π interaction: IQA perspective Phys Chem Chem Phys 2015 Oct 21;17(39):26183-9
M Luck*, A Yartseva, G Bertho, E Thervet, P Beaune, N Pallet, C Damon Metabolic profiling of 1H NMR spectra in Chronic Kidney Disease with local predictive modeling Proc Int Conf Mach Learn Appl, 2015, 176-181
L Ducassou, F André, B Ramassamy, Y Xu-Li, M-A Loriot, P Beaune, G Bertho, M Lombard, D Mansuy, J-L Boucher Expression in Yeast and Discovery of New Substrates of Human Orphan Cytochrome P450 2U1: Interpretation of their Hydroxylation Regioselectivity from Docking Studies on a Protein 3D Model Biochim Biophys Acta 2015 ; 1850(7), 1426-37
2014
N Pallet, E Thervet, P Beaune, A Karras, G Bertho* The urinary metabolome of chronic kidney disease Kidney Int, 2014, 85, 1239–1240
M Carichon, N Pallet, C Schmitt, T Lefebvre, L Gouya, N Talbi, J-C Deybach, P Beaune, P Vasos, H Puy, G Bertho* The urinary metabolic fingerprint of Acute Intermittent Porphyria analyzed by 1H-NMR spectroscopy Anal Chem, 2014, 86, 2166−2174
S Téletchéa, V Stresing, S Hervouet, M Baud’huin, M-F Heymann, G Bertho, C Charrier, K Ando, D Heymann Novel RANK antagonists for the treatment of bone resorptive disease: Theoretical predictions and experimental validation J Bone Miner Res 2014, 29, 1466-1477
A Sadet, L Fernandes, F Kateb, R Balzan, PR Vasos Long-lived coherences: Improved dispersion in the frequency domain using continuous-wave and reduced-power windowed sustaining irradiation J Chem Phys 2014, 141 (5), 054203
2013
F Kateb, H Perrin, K Tripsianes, P Zou, R Spadaccini, M Bottomley, TM Franzmann, J Buchner,S Ansieau, M Sattler Structural and Functional Analysis of the DEAF-1 and BS69 MYND Domains PLoS One 2013, 8: e54715
L Fernandes, C Guerniou, I Marín‐Montesinos, M Pons, F Kateb, PR Vasos Long‐lived states in an intrinsically disordered protein domain Magn Reson Chem 2013, 51 (11), 729-733
C Decroos, V Balland, J-L Boucher, G Bertho, Y Xu, D Mansuy Towards Stable Electron Paramagnetic Resonance Oximetry Probes Synthesis, Characterization and Metabolic Evaluation of New Ester Derivatives of a Tris-(para-carboxyltetrathiaaryl)methyl (TAM) RadicalChem Res Toxicol, 2013, 26, 1561–1569
P Dansette, D Levent, A Hessani, G Bertho, D Mansuy Thiolactone Sulfoxides as New Reactive Metabolites Acting as Bis-Electrophiles: Implication in Clopidogrel and Prasugrel Bioactivation Chem Res Toxicol, 2013, 26, :794-802
C Charrier, G Bertho, O Petigny, P Moneton, R Azerad A new derivative detected in accelerated ageing of artesunate-amodiaquine fixed dose combination tablets J Pharm Biomed Anal 2013, 81-82:20-26
M Hamel, M Lecinq, M Gulea, J Kozelka Ortho-(methylsulfanyl) phenylphosphonates and derivatives: Synthesis and applications as mono-or bidentate ligands for the preparation of platinum complexes J Org Chem, 2013, Vol 745–746, 206–213
Bergès J, Fourré I, Pilmé J, Kozelka J Quantum chemical topology study of the water-platinum(II) interaction Inorg Chem 2013 Feb 4;52(3):1217-27
2012
T Suchánková, K Kubíček, J Kašpárková, V Brabec, J Kozelka Platinum–DNA interstrand crosslinks: Molecular determinants of bending and unwinding of the double helix J Inorg Biochem 2012 Mar;108:69-79
Monnet J, Kozelka J Cisplatin GG-crosslinks within single-stranded DNA: origin of the preference for left-handed helicity J Inorg Biochem 2012 Oct;115:106-12
A Mantsyzov, G Bouvier, N Evrard-Todeschi, G Bertho* Contact-based ligand-clustering approach for the identification of active compounds in virtual screening Adv Appl Bioinform Chem, 2012, doi:102147/AABCS30881
Fougeray, I Mami, G Bertho, C Legendre, P Beaune, E Thervet, N Pallet Tryptophan depletion and the kinase GCN2 mediate interferon γ-induced autophagy J Immunol, 2012, 189, 2954-2964
P Rada, A I Rojo, N Evrard-Todeschi, N G Innamorato, A Cotte, T Jaworski, J C Tobón-Velasco, H Devijver, M Flor García-Mayoral, F Van Leuven, J D Hayes, G Bertho*, A Cuadrado* Structural and functional characterization of Nrf2 degradation by the GSK-3/β-TrCP axis Mol Cell Biol, 2012, 32, 3486-3499
P Dansette, J Rosi, J Debernardi, G Bertho, D Mansuy Metabolic Activation of Prasugrel : Nature of the two Competitive Pathways Resulting in the Opening of its Thiophene RingChem Res Toxicol, 2012, 25, 1058–1065
C Chopard, T Prangé, G Bertho Naphthalene-dioxygenase catalysed cis-dihydroxylation of bicyclic azaarenes RSC Adv, 2012, 2, 605-615
PM Dansette, J Rosi, G Bertho, D Mansuy Cytochromes P450 Catalyze Both Steps of the Major Pathway of Clopidogrel Bioactivation Whereas Paraoxonase Catalyzes the Formation of a Minor Thiol Metabolite Isomer Chem Res Toxicol, 2012, 25, 348-356
2011
Kozelka, J* Evaluation of dissociation constants from competition binding experiments based on the relative binding ratio Anal Biochem, 2011,409, 66‐73
Kumpun, S ; Maria, A ; Crouzet, S ; Evrard‐Todeschi, N ; Girault, JP ; Lafont, R* Ecdysteroids from Chenopodium quinoa Willd, an ancient Andean crop of high nutritional value Food Chem, 2011, 125, 1226‐123
Kumpun, S ; Girault, JP ; Dinan, L ; Blais, C ; Maria, A ; Dauphin‐Villemant, C ; Yingyongnarongkul, B ; Suksamrarn, A ; Lafont, R* The metabolism of 20‐hydroxyecdysone in mice: relevance to pharmacological effects and gene switch applications of ecdysteroids. J Steroid Biochem Mol Biol, 2011, 126, 1‐9
Pons, J ; Tanchou, V ; Girault, JP ; Bertho, G ; Evrard‐Todeschi, N* NMR applications for identifying β‐TrCP protein‐ligand interactions Mini Rev Med Chem, 2011, 11, 283‐297
Sarkar, R ; Ahuja, P *; Vasos, PR ; Bornet, A ; Wagnieres, O ; Bodenhausen, G Long‐lived coherences for line‐narrowing in high‐field NMR Progr Nucl Magn Res Spectrosc, 2011, 59, 83‐9
Segawa, TF *; Bornet, A ; Salvi, N ; Mieville, P ; Vitzthum, V ; Carnevale, D ; Jannin, S ; Caporini, MA ; Ulzega, S ; Vasos, PR ; Rey, M ; Bodenhausen, G* Extending Timescales and Narrowing Linewidths in NMR Chimia, 2011, 65, 652‐655
Bornet, A ; Ahuja, P ; Sarkar, R ; Fernandes, L ; Hadji, S ; Lee, SY ; Haririnia, A ; Fushman, D ; Bodenhausen, G ; Vasos, PR* Long‐lived states to monitor protein unfolding by proton NMR Chem. Phys. Chem., 2011, 12, 2729‐2734
2010
Rizzato, S ; Bergès, J ; Mason, SA ; Albinati, A ; Kozelka, J* Dispersion‐driven hydrogen bonding: predicted hydrogen bond between water and platinum(II) identified by neutron diffraction Angew Chem Int Ed, 2010,49, 7440‐7443
Ahuja, P ; Sarkar, R ; Jannin, S ; Vasos, PR *; Bodenhausen, G Proton hyperpolarisation preserved in long‐lived states Chem Commun, 2010, 46, 8192‐8194
Mamadalieva, NZ ; Janibekov, AA ; Girault, JP ; Lafont, R* Two minor phytoecdysteroids of the plant Silene viridiflora. Nat Prod Commun, 2010, 5, 1579‐1582
Bouvier, G ; Evrard‐Todeschi, N ; Girault, JP ; Bertho, G* Automatic clustering of docking poses in virtual screening process using self‐organising map Bioinformatics, 2010, 26, 53‐60
2009
Kozelka, J* Molecular origin of the sequence‐dependent kinetics of reactions between cisplatin derivatives and DNA Inorg Chim Acta, 2009,362, 651–668
Téletchéa, S ; Skauge, T ; Sletten, E ; Kozelka, J* Cisplatin Adducts on a GGG Sequence within a DNA Duplex Studied by NMR Spectroscopy and Molecular Dynamics Simulations Chem. Eur. J., 2009,15, 12320 – 12337
Crouzet, S ; Maria, A ; Dinan, L ; Lafont, R ; Girault, JP* Ecdysteroids from Cyanotis longifolia Benth. (Commelinaceae). Arch Insect Biochem Physiol, 2009, 72, 194‐209
Zibareva, L ; Yeriomina, VI ; Munkhjargal, N ; Girault, JP ; Dinan, L ; Lafont, R. * The phytoecdysteroid profiles of 7 species of Silene (Caryophyllaceae). Arch Insect Biochem Physiol, 2009, 72, 234‐248
2008
Ho, R ; Girault, JP ; Cousteau, PY ; Bianchini, JP ; Raharivelomanana, P ; Lafont, R* Isolation of a new class of ecdysteroid conjugates (glucosyl‐ferulates) using a combination of liquid chromatographic methods. J Chromatogr Sci, 2008, 46, 102‐110
Bertho, G *;. Bouvier, G ; Hui Bon Hoa, G ; Girault, JP* The key‐role of tyrosine 155 in the mechanism of prion transconformation as highlighted by a study of sheep mutant peptides. Peptides, 2008, 29, 1073‐1084
Pons, J ; Evrard‐Todeschi, N ; Bertho, G ; Gharbi‐Benarous, J ; Tanchou, V ; Benarous, R ; Girault, JP* Transfer‐NMR and Docking Studies Identify the Binding of the Peptide Derived from Activating Transcription Factor 4 to Protein Ubiquitin Ligase beta‐TrCP. Competition STD‐NMR with beta‐Catenin Biochemistry, 2008, 47, 14‐29 (Hot article)
Evrard‐Todeschi, N ; Pons, J ; Gharbi‐Benarous, J ; Bertho, G ; Benarous, R ; Girault, JP* Structure of the Complex between Phosphorylated Substrates and the beta‐TrCP Ubiquitine Ligase Receptor: A combined NMR, Molecular Modelling and Docking Approach J Chem Inf Model, 2008, 48, 2350‐2361
2007
Pons, J ; Evrard‐Todeschi, N ; Bertho, G ; Gharbi‐Benarous, J ; Sonois, V ; Benarous, R ; Girault, JP* Structural studies on 24P‐IkBa peptide derived from a human IkB‐a protein‐related with the inhibition of the transcription factor nuclear NFkB activity Biochemistry, 2007, 46, 2958‐2972
Pons, J ; Evrard‐Todeschi, N ; Bertho, G ; Gharbi‐Benarous, J ; Benarous, R ; Girault, JP* Phosphorylation‐Dependent Structure of ATF4 Peptides derived from a Human ATF4 Protein, a Member of the Family of Transcription Factors Peptides, 2007, 28, 2253‐2267
Snogan, E ; Vahirua‐Lechat, I ; Ho, R ; Bertho, G ; Girault, JP ; Ortiga, S ; Maria, A ; Lafont R* Ecdysteroids from the Medicinal Fern Microsorum scolopendria (Burm. f.) Phytochem Anal., 2007, 18, 441‐450
Aitken, DJ ; Albinati, A ; Gutier, A ; Husson, H‐P ; Morgant, G ; Nguyen‐Huy, D ; Kozelka, J *; Lemoine, P ; Ongeri, S ; Rizzato, S ; Viossat, B Platinum(II) and Palladium(II) Complexes with N‐Aminoguanidine Eur J Inorg Chem, 2007, 3327 ‐ 3334
Hamel, M ; Rizzato, S ; Lecinq, M ; Sene, A ; Vazeux, M ; Gulea, M ; Albinati, A ; Kozelka, J* Study of Intramolecular Competition between Carboxylate and Phosphonate for PtII with the Aid of a Novel Tridentate Carboxylato‐Thioether‐Phosphonato Ligand Chem. Eur. J., 2007,13, 5441‐5449

